This repository contains an implementation of a Multi-output Gaussian processes (MOGPs) model to predict dose response curves and an implementation of a features relevance determination method based on the Kullback-Leibler divergence.
The main experiments are presented in:
- MOGPTraining_and_KLRelevance_Melanoma_GDSC1_and_GDSC2.ipynb, in this notebook:
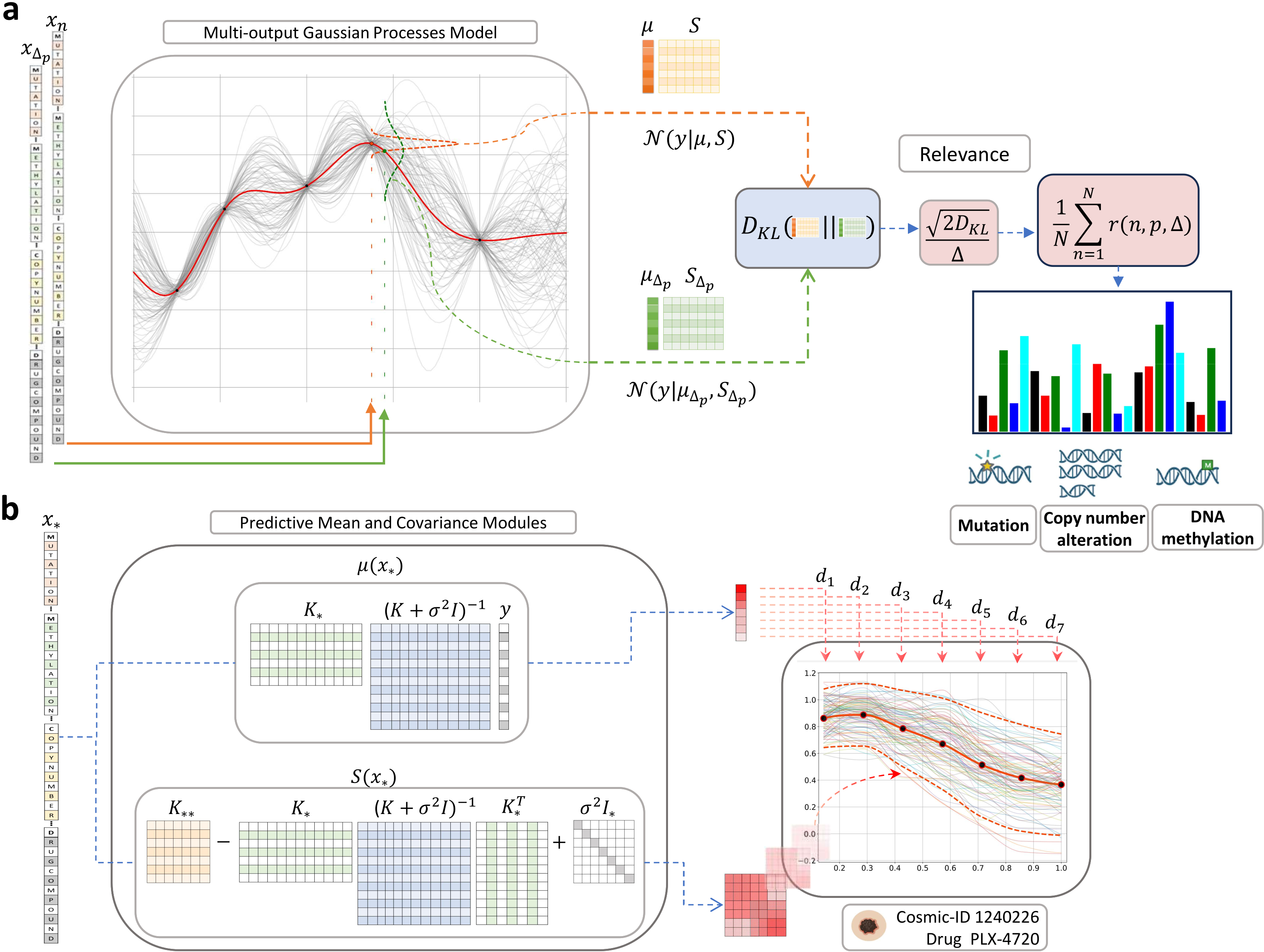
- We train a MOGP model using the GDSC1 dataset and compute a features ranking by means of the KL-Relevance determination method.
- We train the MOGP model using the GDSC2 dataset and compute a features ranking via KL-Relevance method.
- We generate a figure to compare both KL-Relevance rankings obtained with the MOGP models trained on GDSC1 and GDSC2.
- ExactMOGP_TrainOn_MelGDSC1_ToPredict_MelGDSC2_ANOVAFeatures.ipynb, in this notebook:
- We train a MOGP model using the GDSC1 dataset and then assess the performance of the model using the GDSC2 dataset as testing to obtained the summary metrics: IC50, AUC and Emax.
- We generate a plot at the end to compare the fitted reference Sigmoid4_parameters function against the MOGP summary metric predictions.
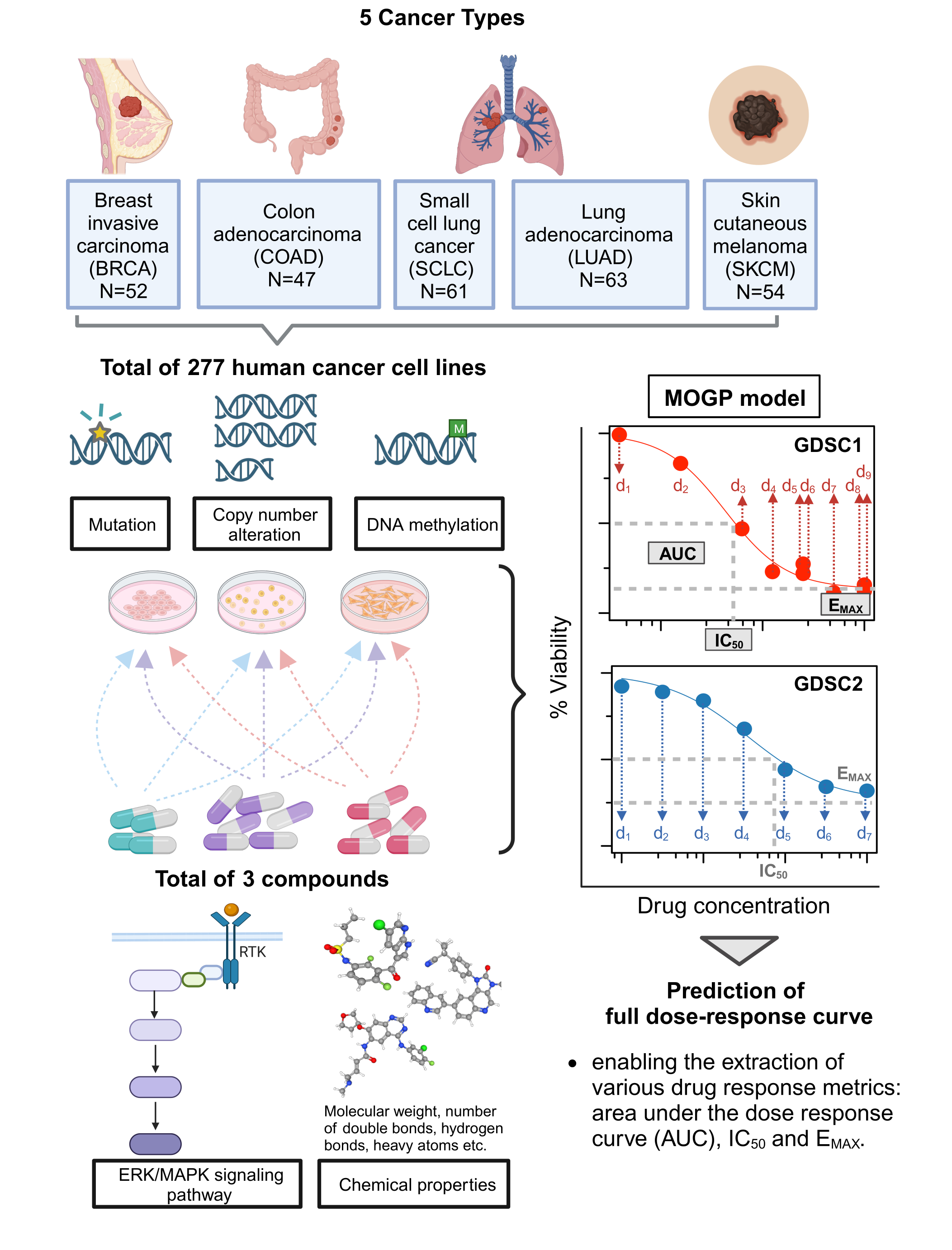
Two datasets, GDSC1 and GDSC2, were constructed by consolidating dose-response data for three drugs (Dabrafenib, PLX-4720, SB590885) targeting the ERK/MAPK pathway, on 277 human cancer cell lines sourced from the GDSC database. Both datasets consist of cancer cell lines representing five different cancer types (BRCA, COAD, SCLC, LUAD and SKCM). Molecular features characterising these cell lines (genetic variations, copy number alterations, DNA methylation) and the chemical properties of the three drugs (sourced from PubChem) were also included. These comprehensive datasets served as input for the MOGP model for predicting full dose-response curves and estimating the relative importance of these input features based on KL divergence.
(a) Kullback-Leibler relevance determination to estimate feature importance. To compute the relevance of a feature w.r.t a data observation we have to make two predictions, one for the original observation