Michael Jahn, David Leslie, Ahmadou Dicko 2021-12-09
Generate and plot Voronoi treemaps or Sunburst treemaps from hierarchical data.
25 March 2021
A Shiny app for generating treemaps from custom data is now available on Shinyapps.io!
Treemaps are a visually appealing graphical representation of numerical data using a space-filling approach. A plane or ‘map’ is subdivided into smaller areas called cells. The cells in the map are scaled according to an underlying metric which allows to grasp the hierarchical organization and relative importance of many objects at once. This package contains two different implementations of treemaps, Voronoi treemaps and Sunburst treemaps
There are different implementations available for Voronoi
tesselations in R, the simplest being the deldir() function (from
package deldir). However, deldir and others do not handle nested
Voronoi tesselations, nor do they perform additively weighted Voronoi
tesselation. This is an important demand for systems biology and other
applications where it is useful to scale the cell size (or area) to a
set of predefined weights. The voronoiTreemap() function provided in
this packages allows both the additively weighted Voronoi tesselation
and the nesting of different hierarchical levels in one plot.
Some of the underlying functions for the tesselation were developed by
Paul Murrell, University of Auckland, and serve as the basis for this
package. They are called by a recursive wrapper function,
voronoiTreemap(), which subdivides the plot area in polygonal cells
according to the highest hierarchical level. It then continues with the
tesselation on the next lower level using the child cell of the previous
level as the new parental cell, and so on.
The Sunburst treemap is a computationally less demanding treemap
that does not require iterative refinement, but simply generates circle
sectors that are sized according to predefined weights. The main
function to draw Sunburst treemaps is sunburstTreemap(). It uses the
same underlying recursive algorithm under the hood and can be used to
draw sectors of different hierarchical levels with increasing
granularity.
The C++ code computing the actual Voronoi tesselation requires the
CGAL library headers. Thanks to
Ahmadou Dicko, installing the complete CGAL
library locally is no longer necessary. Instead, the package depends on
the CGAL4 headers that are now available on CRAN as R package cgal4h.
The dependencies are installed automatically and manual installation of
CGAL (headers) should not be necessary.
To install the package directly from github, use the following function
from the devtools package in your R session:
devtools::install_github("m-jahn/WeightedTreemaps")The functions to create Voronoi (or Sunburst) treemaps take a
data.frame as main input. The data.frame should contain column(s)
with numerical or categorical data (i.e. a character vector). Let’s
create a simple example data frame.
library(WeightedTreemaps)
# load example data
data(mtcars)
mtcars$car_name = gsub(" ", "\n", row.names(mtcars))Generate the treemap. It will return a list of polygons and metadata. The columns of the data frame that are used as levels of the treemap need to be specified. Different parameters like the initial shape, or the maximum number of iterations are optional.
# generate treemap; set seed to obtain same pattern every time
tm <- voronoiTreemap(
data = mtcars,
levels = c("gear", "car_name"),
cell_size = "wt",
shape = "rounded_rect",
seed = 123
)Draw the treemap.
drawTreemap(tm, label_size = 2.5, label_color = "white")The voronoiTreemap() and drawTreemap() functions are separated in
order to allow drawing of the same treemap object in different ways.
Computation of treemaps with thousands of cells can be very time and
resource consuming (around 5-10 minutes for a 2000-cell treemap on a
regular desktop computer). With the drawTreemap() function, we can not
only plot the same treemap in different ways but also combine several
treemaps on one page using the layout and position arguments. The
most important style element is color. Coloring can be based on cell
category, cell size, or both, using the color_type argument. By
default, the highest hierarchical level is used for coloring but that
can be customized using the color_level argument.
drawTreemap(tm, title = "treemap 1", label_size = 2,
color_type = "categorical", color_level = 1,
layout = c(2, 2), position = c(1, 1), legend = TRUE)
#> Note: use 'add = TRUE' if you want to add more treemaps to this page.
drawTreemap(tm, title = "treemap 2", label_size = 2,
color_type = "categorical", color_level = 2, border_size = 3,
add = TRUE, layout = c(2, 2), position = c(1, 2), legend = TRUE)
drawTreemap(tm, title = "treemap 3", label_size = 2,
color_type = "both", color_level = 1,
add = TRUE, layout = c(2, 2), position = c(2, 1), legend = TRUE)
drawTreemap(tm, title = "treemap 4", label_size = 2,
color_type = "cell_size", color_level = 2,
color_palette = heat.colors(10),
border_color = grey(0.4), label_color = grey(0.4),
add = TRUE, layout = c(2, 2), position = c(2, 2),
title_color = "black", legend = TRUE)Generating a Voronoi treemap is an iterative and somewhat random
process. Since the cells ‘move’ during the iteration process, it can be
difficult to control the exact final position of cells. However, there
are two ways to influence cell positioning. The first is to use
different algorithms for sampling initial coordinates for each cell. The
second is simply setting a seed, which will sample the same set of
starting coordinates for the same input data. Regarding the
positioning argument, compare the following three examples where
initial positions are 1) random, 2) ordered from top to bottom, or 3)
ordered from center to edges.
# set seed to obtain same df every time
set.seed(123)
df <- data.frame(A = sample(10:100, 45))
tm1 <- voronoiTreemap(
data = df, levels = "A",
cell_size = "A",
shape = "rounded_rect",
positioning = "random"
)
tm2 <- voronoiTreemap(
data = df, levels = "A",
cell_size = "A",
shape = "rounded_rect",
positioning = "regular"
)
tm3 <- voronoiTreemap(
data = df, levels = "A",
cell_size = "A",
shape = "rounded_rect",
positioning = "clustered"
)drawTreemap(tm1, title = "positioning = 'random'", border_size = 3,
layout = c(1,3), position = c(1, 1))
#> Note: use 'add = TRUE' if you want to add more treemaps to this page.
drawTreemap(tm2, title = "positioning = 'regular'", border_size = 3,
add = TRUE, layout = c(1,3), position = c(1, 2))
drawTreemap(tm3, title = "positioning = 'clustered'", border_size = 3,
add = TRUE, layout = c(1,3), position = c(1, 3))Instead of using predefined shapes, we can also supply a custom set of coordinates to plot a treemap using the `shape``argument. The validity of the supplied coordinates is not checked, so all responsibility lies with the user (!). The R session might even crash (due to C++ dependency) if a shape is supplied that is too irregular or edgy, and the tesselation becomes unfeasible. Here are some stable examples.
# different initial shapes, the more squared the better
house_coords <- list(
x = c(0, 10, 10, 5, 0),
y = c(0, 0, 10,15,10))
rect_coords <- list(
x = c(0, 10, 10, 0),
y = c(0, 0, 3, 3))
oct_coord <- list(
x = sin(seq(0, 2, 2/8)*pi) * 1000 + 1000,
y = cos(seq(0, 2, 2/8)*pi) * 1000 + 1000
)Let’s generate treemaps with the shapes of a house, a rectangle, or an octogon.
tm1 <- voronoiTreemap(data = df, levels = "A",
shape = house_coords)
tm2 <- voronoiTreemap(data = df, levels = "A",
shape = rect_coords)
tm3 <- voronoiTreemap(data = df, levels = "A",
shape = oct_coord)drawTreemap(tm1, layout = c(1,3), position = c(1, 1))
#> Note: use 'add = TRUE' if you want to add more treemaps to this page.
drawTreemap(tm2, add = TRUE, layout = c(1,3), position = c(1, 2))
drawTreemap(tm3, add = TRUE, layout = c(1,3), position = c(1, 3))This example will cover the generation of a somewhat larger treemap, as it is often useful to visualize e.g. many genes or proteins at once in molecular biology studies. However, treemaps can be used for any type of data visualization. First we read a proteomics test data set from Jahn et al., Cell Reports, 2018. (https://www.cell.com/cell-reports/fulltext/S2211-1247(18)31485-2) This dataset contains thousands of protein measurements of the cyanobacterium Synechocystis sp. PCC6803.
# additional libraries for data filtering and colors
library(dplyr)
library(colorspace)
# pick the top most abundant proteins
df <- Jahn_CellReports_2018 %>%
filter(condition == "CO2-0-15") %>%
arrange(desc(mean_mass_fraction_norm)) %>%
slice(1:1000)We can generate the Voronoi treemap using some more of the function’s
parameters. We can increase maxIterations and decrease error_tol
which will lead to lower errors (difference between target cell size and
actual cell size). Set a seed to obtain a similar arrangement of cells
for similar maps, otherwise starting positions will be sampled more
randomly. The positioning argument clustered_by_area will try to
place cells with bigger target area in the middle and smaller area at
the edges.
tm <- voronoiTreemap(
data = df,
levels = c("Process.abbr", "protein"),
cell_size = "mean_mass_fraction_norm",
shape = "rectangle",
error_tol = 0.005,
maxIteration = 200,
positioning = "clustered_by_area",
seed = 1
)Generating and plotting of treemaps are two processes separated on
purpose. Computing treemaps can be time-consuming and to recalculate
them every time just for changing a color gradient or label size is
inefficient. Once a treemap is computed, it can be drawn in different
ways as the following example shows. First we can generate custom color
palettes using colorspaces hclwizard. Just browse to the Export
and then the R tab and copy the code to your script.
# outcomment to run interactive wizard:
#hclwizard()
custom_pal_1 <- sequential_hcl(
n = 20,
h = c(-46, 78),
c = c(61, 78, 54),
l = c(60, 91),
power = c(0.8, 1),
rev = TRUE
)
custom_pal_2 <- diverging_hcl(
n = 7,
h = c(340, 128),
c = c(60, 80),
l = c(75, 97),
power = c(0.8, 1.5),
rev = TRUE
)Draw a customized treemap using some of the graphical parameters. Compare two different color palettes.
drawTreemap(
tm,
color_palette = custom_pal_1,
color_type = "cell_size",
color_level = 2,
label_level = c(1,2),
label_size = 2,
label_color = grey(0.5),
border_color = grey(0.65),
layout = c(1, 2),
position = c(1, 1)
)
drawTreemap(
tm,
color_palette = custom_pal_2,
color_type = "cell_size",
color_level = 2,
label_level = c(1,2),
label_size = 2,
label_color = grey(0.5),
border_color = grey(0.9),
layout = c(1, 2),
position = c(1, 2),
add = TRUE
)This is an example how several treemaps can be computed in parallel.
This functionality is not part of this package but just makes use of
functions contained in the parallel package. First read the test data
set with cyanobacterial proteomics data from 10 different growth
conditions. Only the most abundant proteins are selected for treemap
generation to reduce computation time.
library(parallel)
df <- Jahn_CellReports_2018 %>%
group_by(condition) %>%
arrange(desc(mean_mass_fraction_norm)) %>%
slice(1:200)Generate 10 treemaps using the parallel version of lapply, and the
condition annotation to subset the data frame. Note that you can
adjust the mc.cores parameter to the number of CPUs available on your
computer. The positioning parameter can also take a vector of
length(levels) to make cell positions on the first level more
comparable between different treemaps.
tm <- mclapply(
unique(df$condition),
mc.cores = 10,
mc.set.seed = FALSE,
FUN = function(cond) {
voronoiTreemap(
data = filter(df, condition == cond),
levels = c("Process.abbr", "protein"),
cell_size = "mean_mass_fraction_norm",
custom_color = "mean_mass_fraction_norm",
shape = "rounded_rect",
positioning = c("regular", "clustered_by_area"),
maxIteration = 50,
error_tol = 0.01
)
}
)Draw all 10 treemaps on one canvas using layout and position
arguments.
lapply(1:10, function(i) {
drawTreemap(
tm[[i]],
color_type = "custom_color",
color_level = 2,
color_palette = custom_pal_2,
custom_range = c(0, 0.05),
border_size = 6,
border_color = grey(0.9),
label_level = c(1,2),
label_size = 1.5,
label_color = grey(0.4),
legend = TRUE,
title = unique(df$condition)[i],
title_size = 1.5,
title_color = grey(0.4),
layout = c(2, 5),
position = c(
ifelse(i <= 5, 1, 2),
ifelse(i <= 5, i, i-5)),
add = ifelse(i == 1, FALSE, TRUE)
)
}) %>% invisibleSunburst treemaps are generated in the same way as described above for
Voronoi treemaps. The function to generate a sunburst treemap is
sunburstTreemap(), and just like voronoiTreemap() it returns an
object of class treemapResult (essentially a list) with polygons and
metadata. Drawing is done using the same drawTreemaps() function as
for Voronoi treemaps.
# generate data frame
set.seed(123)
df <- data.frame(
A = rep(c("a", "b", "c"), each = 15),
B = sample(letters[4:12], 45, replace = TRUE)
)
head(df)
#> A B
#> 1 a f
#> 2 a f
#> 3 a e
#> 4 a i
#> 5 a h
#> 6 a gGenerate sunburst treemap.
# by default cell (sector) size is encoded by number of members per group
tm <- sunburstTreemap(
data = df,
levels = c("A", "B")
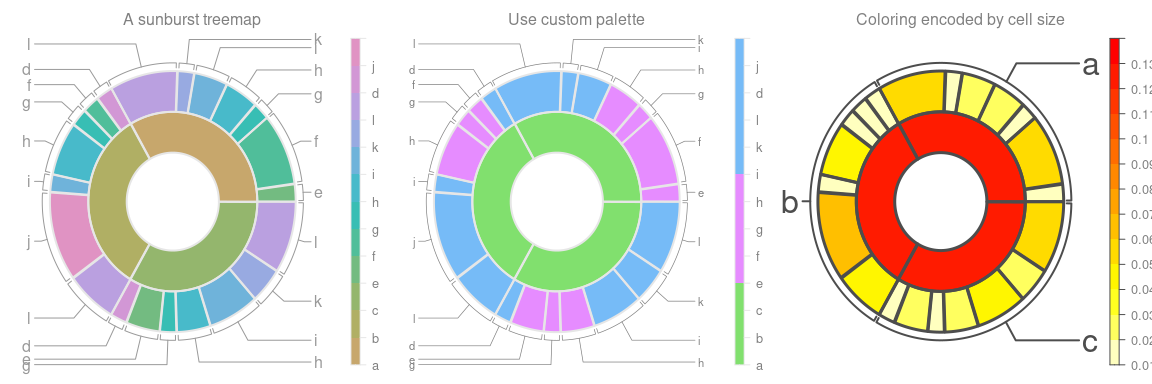
)Draw treemaps with different graphical parameters
# draw treemap with default options
drawTreemap(tm,
title = "A sunburst treemap",
legend = TRUE,
border_size = 2,
label_color = grey(0.6),
layout = c(1, 3),
position = c(1, 1)
)
#> Note: use 'add = TRUE' if you want to add more treemaps to this page.
# use custom color palette
drawTreemap(tm,
title = "Use custom palette",
legend = TRUE,
color_palette = rep(c("#81E06E", "#E68CFF", "#76BBF7"), c(3, 4, 5)),
border_size = 2,
label_level = 2,
label_size = 0.7,
label_color = grey(0.5),
layout = c(1, 3),
position = c(1, 2),
add = TRUE
)
# color cells (sectors) based on cell size
drawTreemap(tm,
title = "Coloring encoded by cell size",
color_type = "cell_size",
legend = TRUE,
color_palette = rev(heat.colors(10)),
border_size = 3,
border_color = grey(0.3),
label_level = 1,
label_size = 2,
label_color = grey(0.3),
layout = c(1, 3),
position = c(1, 3),
add = TRUE
)The Voronoi tesselation is based on functions from Paul Murrell, https://www.stat.auckland.ac.nz/~paul/Reports/VoronoiTreemap/voronoiTreeMap.html. We created a recursive wrapper around the main tesselation function and improved the stability regarding generation of larger treemaps.
For a similar but JAVA based implementation of Voronoi treemaps wrapped in R, see David Leslie’s scripts at https://github.com/dlesl/voronoi_treemap_rJava.
A Javascript based R package lets you draw simple treemaps in your browser, however, this is not suitable for treemaps with many (as, hundreds of) cells. The package is available from CRAN or github, https://github.com/uRosConf/voronoiTreemap.
Another popular resource is the web-based treemap generation from University of Greifswald at https://bionic-vis.biologie.uni-greifswald.de/.