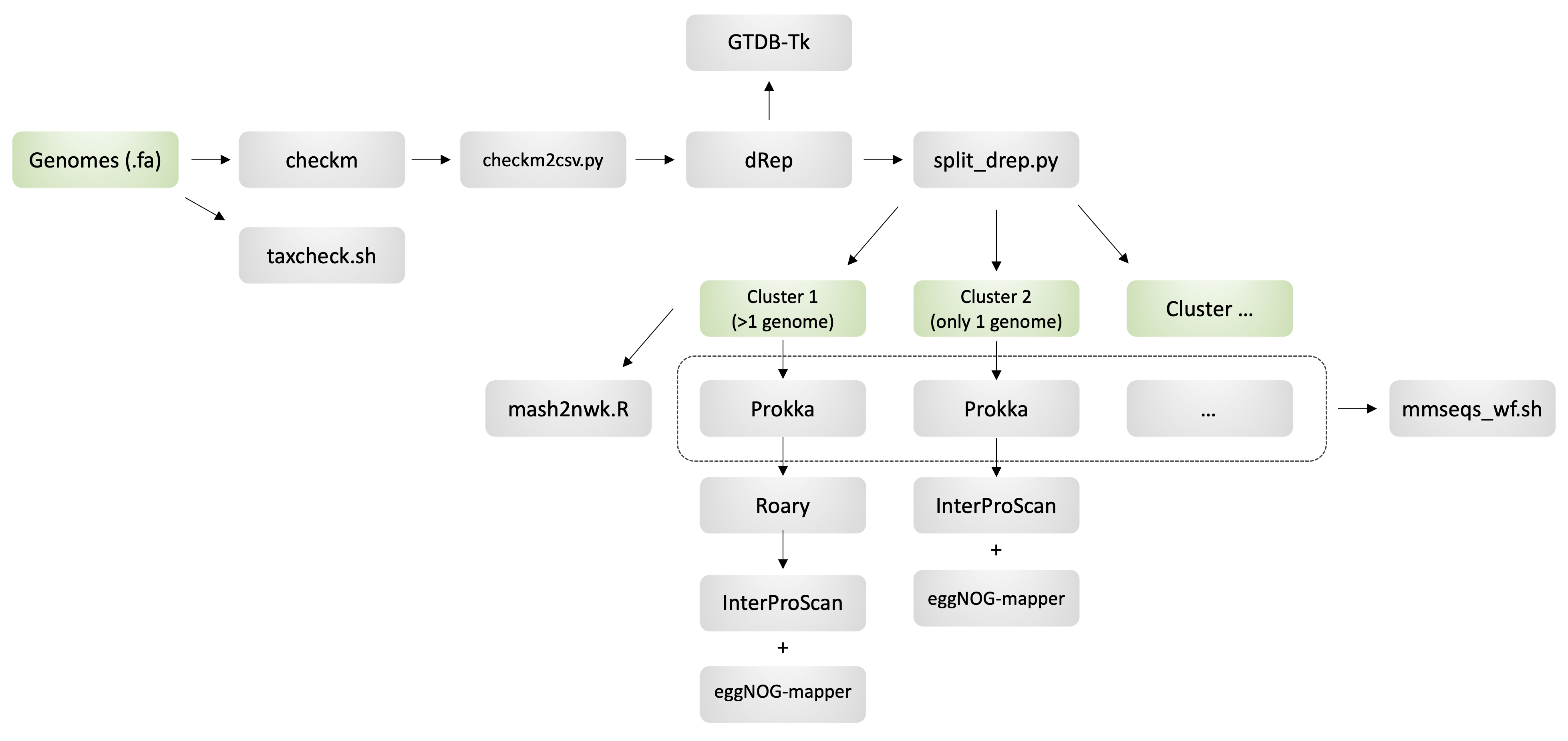
MGnify CWL pipeline to characterize a set of isolate or metagenome-assembled genomes (MAGs) using the workflow described in the following publication:
A Almeida, S Nayfach, M Boland, F Strozzi, M Beracochea, ZJ Shi, KS Pollard, DH Parks, P Hugenholtz, N Segata, NC Kyrpides and RD Finn. A unified sequence catalogue of over 280,000 genomes obtained from the human gut microbiome. bioRxiv. doi: https://doi.org/10.1101/762682
- Install the necessary dependencies:
- cwltool (tested v1.0.2)
- R (tested v3.5.2). Packages: reshape2, fastcluster, optparse, data.table and ape.
- Python v2.7 and v3.6
- CheckM (tested v1.0.11)
- CAT (tested v5.0)
- GTDB-Tk (tested v0.3.1 and v1.0.2)
- dRep (tested v2.2.4)
- Prokka (tested 1.14.0)
- Roary (tested 3.12.0)
- MMseqs2 (tested v8-fac81)
- InterProScan (tested v5.35-74.0 and v5.38-76.0)
- eggNOG-mapper (tested v2.0)
-
Make sure all these tools, as well as the custom_scripts/ folder, are added to your
$PATHenvironment. -
Edit custom_scripts/taxcheck.sh to point CAT to the installed diamond and database paths (variables
$diamond_path,$cat_db_pathand$cat_tax_path)
-
Add path of input genomes folder to YML file: workflows/yml_patterns/wf-1.yml
-
Run first workflow with:
cwltool workflows/wf-1.cwl workflows/yml_patterns/wf-1.yml > output-wf-1.json
Output json will be saved to a separate file. -
Run parser of output json
python3 workflows/parser_yml.py -j output-wf-1.json -y workflows/yml_patterns/wf-2.yml -
Check exit code of parser
echo $? -
If exit code == 1, run:
cwltool workflows/wf-exit-1.cwl workflows/yml_patterns/wf-2.yml
If exit code == 2, run:
cwltool workflows/wf-exit-2.cwl workflows/yml_patterns/wf-2.yml
If exit code == 3, run:
cwltool workflows/wf-exit-3.cwl workflows/yml_patterns/wf-2.yml
Note: You can manually change parameters of MMseqs2 for protein clustering in workflows/yml_patterns/wf-2.yml
Output files/folders:
- checkm_quality.csv
- gtdb-tk_output/
- taxcheck_output/
- mmseqs_output/
- mash_trees/
- cluster__X
- cluster__...
- CheckM: Estimate genome completeness and contamination.
- TaxCheck: Wrapper of the contig annotation tool (CAT) to predict taxonomy consistency across contigs.
- GTDB-Tk: Genome taxonomic assignment using the GTDB framework.
- dRep: Genome de-replication.
- Mash2Nwk: Generate Mash distance tree of conspecific genomes.
- Prokka: Predict protein-coding sequences from genome assembly.
- Roary: Infer pan-genome from a set of conspecific genomes.
- MMseqs2: Cluster protein-coding sequences.
- InterProScan: Protein functional annotation using the InterPro database.
- eggNOG-mapper: Protein functional annotation using the eggNOG database.
1.1) checkm
1.2) checkm2csv
1.3) dRep
1.4.1) GTDB-Tk
1.4.2) split_drep.py
1.5) classify_folders.py
2) taxcheck
output:
- checkm_csv
- gtdbtk folder
- taxcheck_dirs
plus folders for the next step - one_genome (list of clusters/folders that have only one genome)
- many_genomes (list of clusters/folders that have more than one genome)
- mash_folder (list of mash-files from "many_genomes" clusters)
Check
======> if many_genomes and one_genome present: run wf-exit-1.cwl
**2.1. For many_genomes part**
1) Prokka
2) Roary
3) translate from fa to faa
4.1) IPS
4.2) EggNOG
output: OUTPUT_MANY
- mash_trees
- cluster folder(-s)
- prokka concatenated faa result
**2.2. For one_genome part**
1) Prokka
2.1) IPS
2.2) EggNOG
output: OUTPUT_ONE
- cluster folder(-s)
- prokka concatenated faa result
**2.3. Final part**
1) cat prokka from many and one
2) mmseqs
output: OUTPUT_3
- mmseqs folder
======> if many_genomes present BUT one_genome NOT present: run wf-exit-2.cwl
Step 2.1 + 2.3
======> if many_genomes NOT present BUT one_genome present: run wf-exit-3.cwl
Step 2.2 + 2.3
Copies all relevant output to one result folder