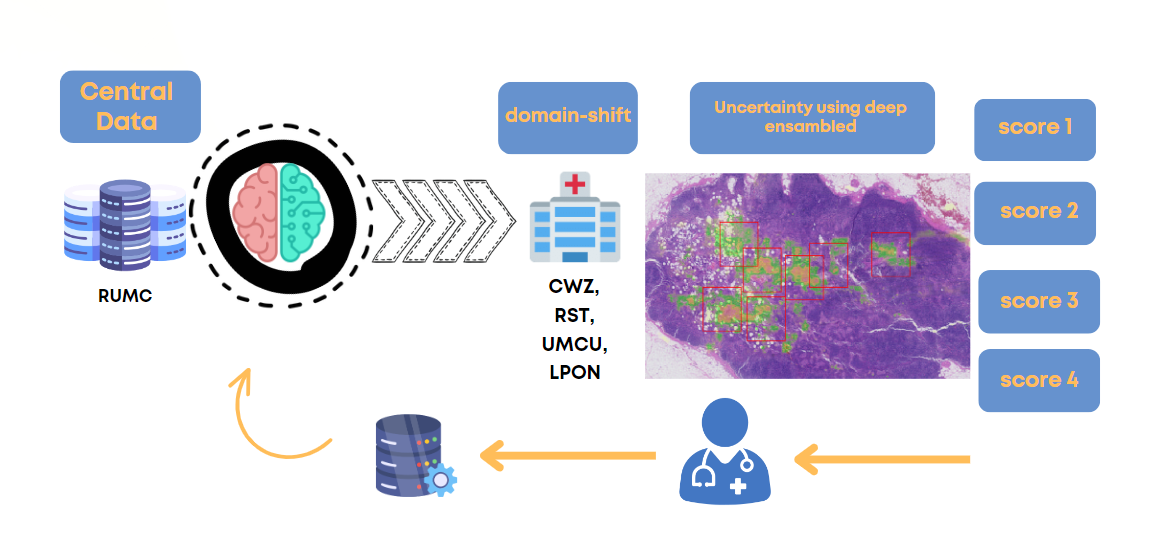
This project introduces a novel human-in-the-loop deep learning pipeline for enhancing the efficiency and accuracy of cancer cell segmentation in digital pathology.
-
Efficiency and Accuracy: Utilizes per-pixel and local-level model uncertainty to guide the annotation process.
-
Informed Decision-making: Provides pathologists with valuable insights into the model's limitations.
-
Out-of-Domain Handling: Specifically designed to handle Out-of-Domain (OOD) scenarios commonly encountered in medical data.
Our study employed the Camelyon dataset, focusing on Whole Slide Images (WSIs) to perform pixel-level segmentation. Training Data: Our model was trained on Camelyon 16, specifically using data obtained from Radboud University Medical Center. Uncertainty Measurement: To evaluate the model's uncertainty, the model was tested on Camyelyon 17 including five centers
Our pipeline demonstrates that using fewer annotated samples can still yield higher segmentation performance. This is achieved by focusing on out-of-domain areas identified through model uncertainty measurements.
The pipeline employs five ensemble models of an adapted version of nnUNet, specifically tuned for pathology data for both segmentation and uncertainty measurements.
- Data: Trained on Whole Slide Images (WSIs).
- Uncertainty Measurements: Incorporates uncertainty to identify and correct misclassifications.
Note: This pipeline is flexible; you can substitute nnUNet with any other network that provides uncertainty measurements.
-
Prepare your dataset:
python3 create_training.py
-
Train the ensembles of the segmentation network: Before proceeding, ensure nnUNet is set up by following the instructions in this repository: nnUNet for Pathology
nnunet plan_train task_name root --network 2d --planner3d None --planner2d ExperimentPlanner2D_v21_RGB_scaleTo_0_1_bs8_ps512 --plans nnUNet_RGB_scaleTo_0_1_bs8_ps512 --trainer nnUNetTrainerV2_BN --fold 0 to 4
-
Compute uncertainty on new domain:
python3 nnunet_inference.py folder taskname ``` -
Create data from the area that the model is the most uncertain about:
python3 uncertainty_csv.py python3 uncertainty_sampling.py
-
Retrain the segmentation network with in-domain and out-of-domain samples:
nnunet plan_train task_name root --network 2d --planner3d None --planner2d ExperimentPlanner2D_v21_RGB_scaleTo_0_1_bs8_ps512 --plans nnUNet_RGB_scaleTo_0_1_bs8_ps512 --trainer nnUNetTrainerV2_BN --fold 0- Test:
python3 nnunet_inference.py folder taskname