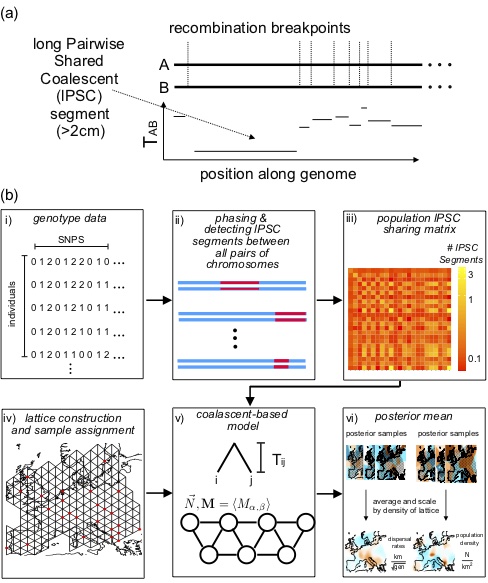
MAPS estimates time-resolved dispersal rates and population densities based on the number of shared long Pairwise Coalescent Segments (lPSC).
MAPS has been tested with Eigen 3.2.2 and Boost 1.57 on OSX and Linux and might not be compatible with newer versions of Boost/Eigen.
Please post bugs, questions and feature requests or suspected bugs to Github issues.
Follow these steps to quickly get started using MAPS.
For installing the software dependencies, we provide detailed instructions using the conda package manager. This is only a recommended aproach---conda is not required to use MAPS, and other approaches can be taken to install the dependencies (e.g., by directly downloading the source code, or by using Homebrew for Mac).
-
Install GNU Make.
-
Install a Conda distribution such as Anaconda or Miniconda (optional).
-
Create a new conda environment for MAPS (optional).
conda create -n MAPS source activate MAPS -
Install the Boost C++ source libraries version 1.57.0. If using conda, run
conda install boost=1.57.0. -
Install the Eigen C++ matrix algebra library. If using conda, run
conda install eigen. -
Clone or download this repository.
-
Adjust the Makefile variables as needed to suit your computing environment. See the instructions in the Makefile for details. For example, if you installed the Boost and Eigen libraries with conda, then you would add the following flags to the
CXXFLAGSandLDFLAGSsettings, respectively (again, see the Makefile for full details):-I${HOME}/anaconda/envs/MAPS/include -L${HOME}/anaconda/envs/MAPS/lib -
Run
makein the src subdirectory to build theruneems2binary executable. -
Make sure that the Boost library is in the library search path. Typically this will involve updating the
LD_LIBRARY_PATH(Linux) orDYLD_LIBRARY_PATH(macOS) environment variable; e.g.,export LD_LIBRARY_PATH=$HOME/anaconda/envs/MAPS/lib
-
Try running MAPS on the 2-6 cM PSC segments provided in the examples/2_6 subdirectory.
cd ../examples ../src/runeems2 --params params-test-2_6.iniIt may take 10--30 minutes for the MCMC simulation to complete.
-
Next, try running MAPS on larger PSC segments (>6 cM) to investigate at more recent population structure.
../src/runeems2 --params params-test-6_Inf.ini
-
install the plotmaps package in R, and run the following code in R. This will generate plots for the MAPS analyses. The plots are saved as PDFs.
library(plotmaps) plot_maps(add.pts = TRUE, add.graph = TRUE, add.countries = FALSE, longlat = TRUE, mcmcpath = "2_6/2_6-MAPS-test-sim", outpath = "2_6", width = 10, height = 6) plot_maps(add.pts = TRUE, add.graph = TRUE, add.countries = FALSE, longlat = TRUE, mcmcpath = "6_Inf/6_Inf-MAPS-test-sim", outpath = "6_Inf", width = 10, height = 6, plot.difference=TRUE)
Continue reading for more detailed documentation on MAPS.
MAPS requires for input
- IBD sharing matrix (ends in .sims) (see below)
- coordinate file (ends in .coord). see eems documentation
- outer file (ends in .outer). see eems documentation
An IBD sharing matrix is required for MAPs (instead of an
disssimilarity matrix as in eems). The IBD sharing matrix popressard_2_Inf.sims.
The length bin R is defined by a lowerbound and an upperbound on the
cM scale, and can be specified in the params file withe the
parameter lowerBound and upperBound. If upperBound entry is
blank, it is assumed to be infinity.
For example,
datapath = popressard_2_Inf.sims
lowerBound = 2
We also allow the capability to visualize IBD segments in a length
region, for example betweem 2cM and 8cM. datapath = popressard_2_8.sims lowerBound = 2 upperBound = 8
The MAPs software is built from the eems software and the software usage is very similar. Here, we only highlight the differences between the usage of MAPs and EEMS. Please see https://github.com/dipetkov/eems for the usage in EEMS.
In comparison to EEMS, there are a few additional arguments in the params file,
-
genomeSize(optional defaults to 3000cM) -
nIndiv(required, number of haploid individuals) -
lowerBound(required, lower bound in cM) -
upperBound(optional defaults to infinity, upper bound in cM) -
olderpath(optional, path to a run with a older time period, MAPS will only estimate the difference between rates from the older time period)
In our paper, we use BEAGLE (https://faculty.washington.edu/browning/beagle/beagle.html) and we make available our pipeline for calling IBD segments: https://github.com/halasadi/ibd_data_pipeline
Also, see the folder convert_beagle for an example on how to convert BEAGLE output to a .sims file.
Note: We provide example scripts, please feel to use any IBD calling software/pipelines you see fit.
If you find the MAPS program, or any source code contained in this repository, useful for your work, please cite:
https://journals.plos.org/plosgenetics/article?id=10.1371/journal.pgen.1007908
Copyright (c) 2017-2018, Hussein Al-Asadi.
The software and example programs in this repository are made available under the terms of the MIT license. See LICENSE for the full text of the license.
This project was developed by Hussein Al-Asadi at the University of Chicago.
Thanks to Matthew Stephens and John Novembre for their support and mentorship.