This repository includes a MATLAB toolbox for remote sensing change detection wherein several classical methods are implemented. I'm working on building an open-source, end-to-end, and extensible benchmark framework for the convenience of the RSCD research.
Generally, a typical RSCD process can be divided into four steps, namely image pre-processing, change detection, binarization (thresholding) and accuracy evaluation. Currently, the toolbox is mainly based on the final three steps. For ease of usage and maintenance, the toolbox adopted modular design, which was realized by packages and classes in MATLAB semantics.
- The
Datasetspackage holds several classes of change detection datasets. Algorithmscollects the change detection methods to yield a difference image.ThreAlgscontains 3 common methods for binary segmentation.Metricsgroups 6 metrics that are frequently used in researches.
Each of these packages contains replaceable modules and the modules from different packages can be freely combined. It is worth mentioning that the modules for pre-processing and post-processing are still under development.
The toolbox is developed and tested in MATLAB R2017b.
For a quick start, use detectChange.m. Here are some examples.
[CM, DI] = detectChange('CVA', {}, 'KMeans', {}, 'data/2000TM', 'data/2003TM')
[CMs, DIs, results] = detectChange('MAD', {}, 'OTSU', {}, '2000TM.tif', '2003TM.tif', 'gt.png', {'AUC', 'UA'}, {{}, {}})See the usage of detectChange.m by
help detectChangeTo try full functionality, find the main script main.m in the root directory of this repo, and follow the three steps below:
First, check and set the global configurations. They are in the beginning of the script and most of them are about logging and visualization settings. So feel free to fiddle. See the comment to understand what an option will do.
Second, select the modules you would like to use by specifying their names in quotes. The names have to be valid and exact, which means that a name ought to be taken from the Name column of the Available Lists. For the METRICS, put any number of items in a cell array. An example is here:
ALG = 'MAD'
DATASET = 'TaizhouDataset'
THRE_ALG = 'KMeans'
METRICS = {'OA', 'UA', 'Recall', 'FMeasure', 'AUC', 'Kappa'}Then, type the arguments by assigning cell arrays to CONFIG_*. The assignment statements have to be there even if no argument is expected. Otherwise, it wil raise an error. For the zero-argument case, just leave the cell empty. Note that for CONFIG_DATASET, a path string that directs to the root directory of the dataset is always required. For CONFIG_METRICS, the arguments of each CDMetric module should be specified in separated sub-cells. And the number of the nested cells matches the length of MRTRICS. See this example:
CONFIG_ALG = {1, 2};
CONFIG_DATASET = {'E:\\Mydataset'};
CONFIG_THRE_ALG = {};
% MRTRICS = {'A', 'B', 'C'}
CONFIG_METRICS = {{'a'}, {}, {}};Run the code 🚀
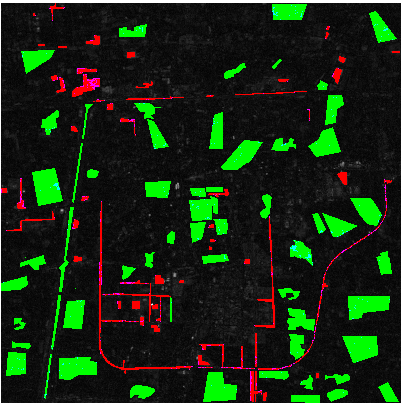
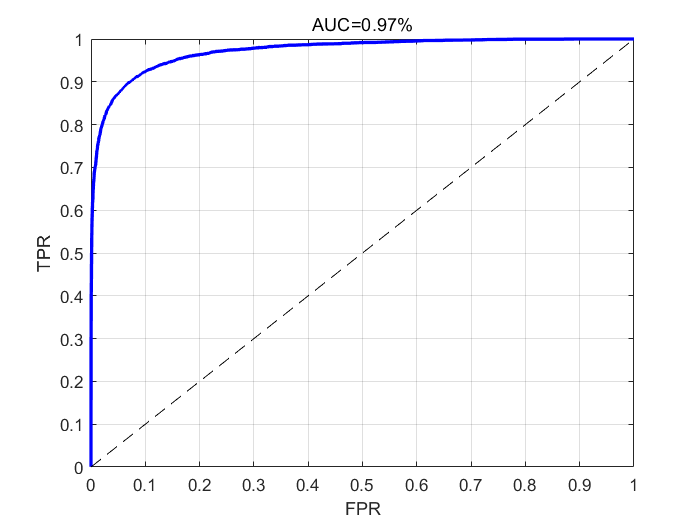
Here are some visualized results
| Dataset | Name | Link |
|---|---|---|
| SZTAKI AirChange Benchmark set | AirChangeDataset | link |
| Bern dataset | BernDataset | |
| Onera Satellite Change Detection dataset | OSCDDataset | link |
| Ottawa dataset | OttawaDataset | |
| Taizhou dataset | TaizhouDataset |
| Algorithm | Name | Paper |
|---|---|---|
| Change Vector Analysis | CVA | paper |
| Differential Principal Component Analysis | DPCA | paper |
| Image differencing | ImageDiff | paper |
| Image ratioing | ImageRatio | paper |
| Iteratively Reweighted MAD | IRMAD | paper |
| Multivariate Alteration Detection | MAD | paper |
| PCA k-Means | PCAkMeans | paper |
| Principal Component Differential Analysis | PCDA | paper |
| Algorithm | Name |
|---|---|
| Using a fixed threshold | FixedThre |
| k-Means clustering | KMeans |
| Otsu's method | OTSU |
| Metric | Name |
|---|---|
| Area under the curve | AUC |
| F-Measure | FMeasure |
| Cohen's kappa coefficient | Kappa |
| Overall accuracy | OA |
| Recall rate | Recall |
| User accuracy | UA |
Small yet useful scripts are provided in the Scripts folder. For example, Scripts/raster2tiff.py would be a nice helper to convert various types of raster images into .tiff format, which is of particular use when transforming a dataset.
This toolbox is designed in modules such that you can easily extend it for a specific task. This can be achieved by class inheritance, and the base classes are Datasets.CDDataset, Metrics.CDMetric, ThreAlgs.ThreAlg, and Algorithms.CDAlg, of which the class names are clear enough to show the functionality. Details should be depicted in the Dev Guide, if there were one.
Thanks to the awesome repo of wenhwu, I couldn't have finished my experiments without these carefully collected datasets. A considerable portion of this work actually referred to the open source community, for which I'd like to thank all these authors. Also, I would like to thank Xie Yachao and Xia Yu for their kind help and useful advice.
This repo is mostly based on the "Anti 996" License and the scripts of reading ENVI files, +Datasets/+Loaders/private/envidataread.m and +Datasets/+Loaders/private/envihdrread.m, are under the MIT license.
- 2019.11.8 Now it is possible to run the program on a single pair of images.
Contributions and suggestions are highly welcomed. Let's work together!