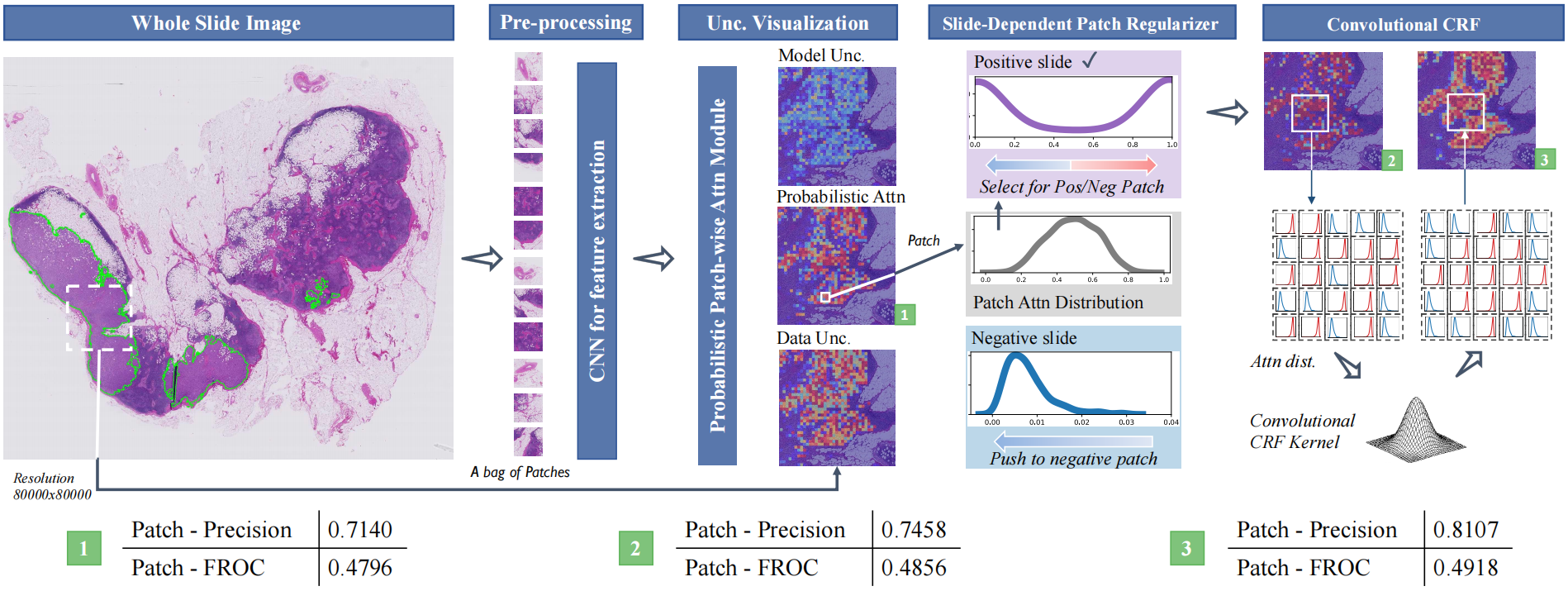
Bayes-MIL: A New Probabilistic Perspective on Attention-based Multiple Instance Learning for Whole Slide Images
This repository is the implementation of Bayes-MIL: A New Probabilistic Perspective on Attention-based Multiple Instance Learning for Whole Slide Images. Parts of this project use code from CLAM.
Use the environment configuration the same as CLAM.
- Create patches
python create_patches_fp.py --source DIR_TO_SLIDE_FILE --save_dir SAVE_DIR --patch_size 256 --seg --patch --stitch- Extract patch features
CUDA_VISIBLE_DEVICES=0,1 python extract_features_fp.py --data_h5_dir DIR_TO_COORDS --data_slide_dir DATA_DIRECTORY --csv_path CSV_FILE_NAME --feat_dir FEATURES_DIRECTORY --batch_size 512 --slide_ext .svs-
Prepare the data in the
dataset_csvfolder.Datasets are expected to be prepared in a csv format containing at least 3 columns: case_id, slide_id, and labels columns for the slide-level labels.
-
Split the data into k-fold(e.g. 10-fold), then save the splited data in the following format as in
splits/x/splits_x.csv.
python create_splits_seq.py --task task_1_tumor_vs_normal --seed 1 --label_frac 0.75 --k 10- Calculate the shape of images.
Create a images_shape.txt file that stored with slide name, width and height of each slide at level 0.
python get_image_shape.py- Modify the format of the input data.
For data loading, look under datasets/dataset_generic.py:
def __getitem__(self, idx):
slide_id = self.slide_data['slide_id'][idx]
label = self.slide_data['label'][idx]
if type(self.data_dir) == dict:
source = self.slide_data['source'][idx]
data_dir = self.data_dir[source]
else:
data_dir = self.data_dir
full_path = os.path.join(data_dir, 'h5_files', '{}.h5'.format(slide_id))
with h5py.File(full_path, 'r') as hdf5_file:
features = hdf5_file['features'][:]
coords = hdf5_file['coords'][:]
features = torch.from_numpy(features)
coords = torch.from_numpy(coords)
w, h = self.shape_dict[slide_id]
return slide_id, features, label, coords, w, hslide_id: Name of slide.features: Features of all patches in the slide.label: Label of slide (0 or 1)coords: The top-left coordinates of all patches in the slide.w: Width of the slide.h: Height of the slide.
- Bayes-MIL.
We consider 3 variants of Bayes-MIL: 1) Bayes-MIL-Vis: The Bayesian modelling of MIL. 2) Bayes-MIL-SDPR: The model with slide-dependent patch regularizer. 3) Bayes-MIL-APCRF: The whole model, including SDPR and the approximate CRF.
For training, look under models/model_bmil.py:
bMIL_model_dict = {
'vis': probabilistic_MIL_Bayes_vis,
'enc': probabilistic_MIL_Bayes_enc,
'spvis': probabilistic_MIL_Bayes_spvis,
}📋 Update
--model_typeto the corresponding model.
bmil-vis: Bayes-MIL-Visbmil-enc: Bayes-MIL-SDPRbmil-spvis: Bayes-MIL-APCRF
CUDA_VISIBLE_DEVICES=0 nohup python -u main.py --drop_out --early_stopping --lr 1e-4 --k 10 --label_frac 0.75 --exp_code task_1_tumor_vs_normal_CLAM_75 --weighted_sample --bag_loss ce --inst_loss svm --task task_1_tumor_vs_normal --model_type bmil-vis --log_data --data_root_dir FEATURES_DIRECTORY &By adding your own custom datasets into eval.py the same way as you do for main.py. Modify the --model_type to the corresponding model.
CUDA_VISIBLE_DEVICES=0 python eval.py --drop_out --k 10 --models_exp_code task_1_tumor_vs_normal_CLAM_75 --save_exp_code task_1_tumor_vs_normal_CLAM_75 --task task_1_tumor_vs_normal --model_type bmil-vis --results_dir results --data_root_dir DATA_ROOT_DIRModify the heatmaps/configs/config_template.yaml to filling out the config. The heatmaps will be saved in heatmaps/heatmap_raw_results.
CUDA_VISIBLE_DEVICES=0 nohup python -u create_heatmaps_bmil.py --config config_template.yaml &This code is made available under the GPLv3 License and is available for non-commercial academic purposes.
Please cite our paper if you use the core code of Bayes-MIL.
Yufei, Cui, et al. "Bayes-MIL: A New Probabilistic Perspective on Attention-based Multiple Instance Learning for Whole Slide Images." The Eleventh International Conference on Learning Representations. 2023.
@inproceedings{yufei2023bayes,
title={Bayes-MIL: A New Probabilistic Perspective on Attention-based Multiple Instance Learning for Whole Slide Images},
author={Yufei, Cui and Liu, Ziquan and Liu, Xiangyu and Liu, Xue and Wang, Cong and Kuo, Tei-Wei and Xue, Chun Jason and Chan, Antoni B},
booktitle={The Eleventh International Conference on Learning Representations},
year={2023}
}