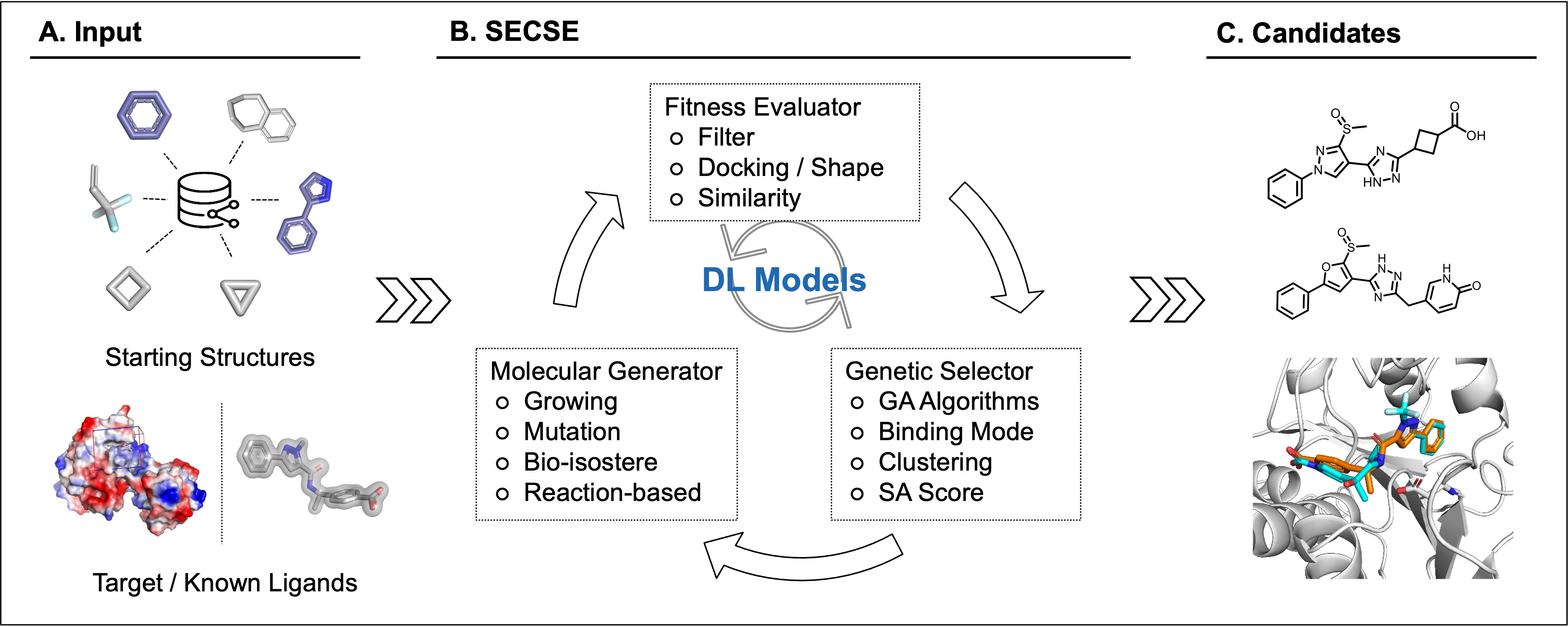
Chemical space exploration is a major task of the hit-finding process during the pursuit of novel chemical entities. Compared with other screening technologies, computational de novo design has become a popular approach to overcome the limitation of current chemical libraries. Here, we reported a de novo design platform named systemic evolutionary chemical space explorer (SECSE). The platform was conceptually inspired by fragment-based drug design, that miniaturized a “lego-building” process within the pocket of a certain target. The key to virtual hits generation was then turned into a computational search problem. To enhance search and optimization, human intelligence and deep learning were integrated. SECSE has the potential in finding novel and diverse small molecules that are attractive starting points for further validation.
-
Set Environment Variables
export SECSE=/path/to/SECSE
if you use AutoDock Vina for docking: (download here)
export VINA=/path/to/AutoDockVINA
if AutoDock GPU: (adgpu-v1.5.3_linux_ocl_128wi) (download here)
export AUTODOCK_GPU=/path/to/AutoDockGPU
if you use Gilde for docking (additional installation & license required):
export SCHRODINGER=/path/to/SCHRODINGER -
Give execution permissions to the SECSE directory
chmod -R +x /path/to/SECSE -
Input fragments: a tab separated .smi file without header. See demo here.
-
Parameters in config file:
[DEFAULT]
- workdir, working directory, create if not exists, otherwise overwrite, type=str
- fragments, file path to seed fragments, smi format, type=str
- num_gen, number of generations, type=int
- num_per_gen, number of molecules generated each generation, type=int
- seed_per_gen, number of selected seed molecules per generation, default=1000, type=int
- start_gen, number of staring generation, default=0, type=int
- docking_program, name of docking program, AutoDock-Vina (input vina) or AutoDock-GPU (input autodock-gpu) or Glide (input glide) , default=vina, type=str
- cpu, number of max invoke CPUs, type=int
- gpu, number of max invoke GPU for AutoDock GPU, type=int
- rule_db, path to customized rule in json format, input 0 if use default rule, default=0
[docking]
- target, protein PDBQT if use AutoDock Vina; grid map files descriptor fld file if AutoDock GPU; Grid file if choose Glide, type=str
- RMSD, docking pose RMSD cutoff between children and parent, default=2, type=float
- delta_score, decreased docking score cutoff between children and parent, default=-1.0, type=float
- score_cutoff, default=-9, type=float
Parameters when docking by AutoDock Vina:
- x, Docking box x, type=float
- y, Docking box y, type=float
- z, Docking box z, type=float
- box_size_x, Docking box size x, default=20, type=float
- box_size_y, Docking box size y, default=20, type=float
- box_size_z, Docking box size z, default=20, type=float
[deep learning]
- mode, mode of deep learning modeling, 0: not use, 1: modeling per generation, 2: modeling overall after all the generation, default=0, type=int
- dl_per_gen, top N predicted molecules for docking, default=100, type=int
- dl_score_cutoff, default=-9, type=float
[properties]
- MW, molecular weights cutoff, default=450, type=int
- logP_lower, minimum of logP, default=0.5, type=float
- logP_upper, maximum of logP, default=7, type=float
- chiral_center, maximum of chiral center,default=3, type=int
- heteroatom_ratio, maximum of heteroatom ratio, default=0.35, type=float
- rotatable_bound_num, maximum of rotatable bound, default=5, type=int
- rigid_body_num, default=2, type=int
Config file of a demo case phgdh_demo_vina.ini
Customized rule json template rules.json. Rule ID should be in the form G-001-XXXX, like G-001-0001, G-001-0002, G-001-0003 ... -
Run SECSE
python $SECSE/run_secse.py --config path/to/config -
Output files
- merged_docked_best_timestamp_with_grow_path.csv: selected molecules and growing path
- selected.sdf: 3D conformers of all selected molecules
GNU Parallel installation
- CentOS / RHEL
sudo yum install parallel - Ubuntu / Debian
sudo apt-get install parallel - From source: https://www.gnu.org/software/parallel/
numpy~=1.20.3, pandas~=1.3.3, pandarallel~=1.5.2, tqdm~=4.62.2, biopandas~=0.2.9, openbabel~=3.1.1, rdkit~=2021.03.5, chemprop~=1.3.1, torch~=1.9.0+cu111
Linux server with CPUs only also works.
Lu, C.; Liu, S.; Shi, W.; Yu, J.; Zhou, Z.; Zhang, X.; Lu, X.; Cai, F.; Xia, N.; Wang, Y. Systemic Evolutionary Chemical Space Exploration For Drug Discovery. J Cheminform 14, 19 (2022).
https://doi.org/10.1186/s13321-022-00598-4
SECSE is released under Apache License, Version 2.0.
Contact email: [email protected]