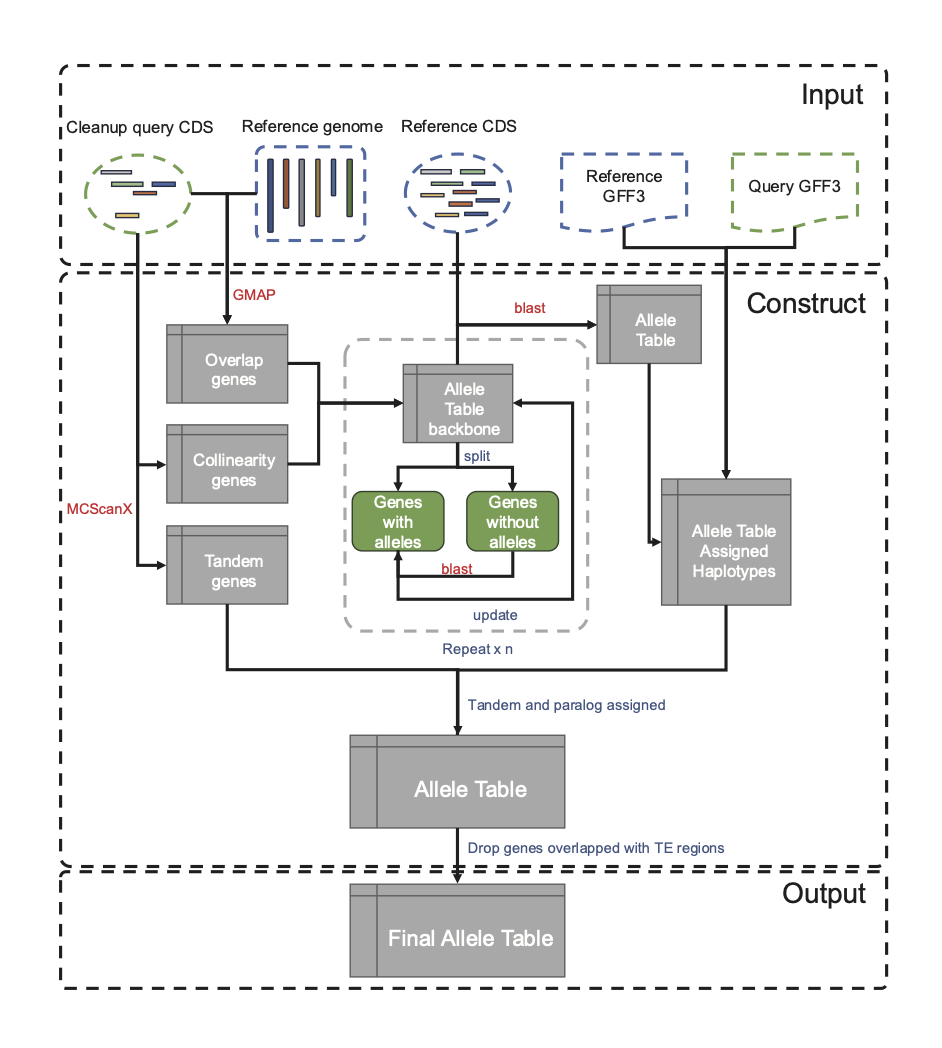
This software is used for identifying allele genes from polyploid genome.
Software:
Notice: these software should be added to the PATH Environment Variable.
cd /path/to/install
git clone https://github.com/sc-zhang/AlleleFinder.git
chmod +x AlleleFinder/allelefinder.py
# Optional
echo 'export PATH=/path/to/install/AlleleFinder:$PATH' >> ~/.bash_profile
source ~/.bash_profileusage: allelefinder.py [-h] {construct,cleanup,stat,adjust} ...
options:
-h, --help show this help message and exit
Sub commands:
{construct,cleanup,stat,adjust}
construct Construct allele table
cleanup Remove same CDS from cds, pep and gff3 files
stat Statistic allele table
adjust Adjust allele table with too many genes be marked as paralogusage: allelefinder.py construct [-h] -r REF -d REF_CDS -f REF_GFF3 -c CDS -g GFF3 -n NUM_ALLELE [-m] [--ovlp_ratio OVLP_RATIO] [-b BLAST_COUNT] [--blast_identity BLAST_IDENTITY] [-e TE] [-j TE_OVERLAP] [--paralog_only] [-w WORKDIR] [-t THREADS]
options:
-h, --help show this help message and exit
-r REF, --ref REF reference fasta
-d REF_CDS, --ref_cds REF_CDS
CDS fasta of ref
-f REF_GFF3, --ref_gff3 REF_GFF3
GFF3 file of ref
-c CDS, --cds CDS CDS fasta of polyploid
-g GFF3, --gff3 GFF3 GFF3 file of polyploid
-n NUM_ALLELE, --num_allele NUM_ALLELE
number of allele
-m, --is_mono if your reference fasta is mono assembly of polyploid, add this argument
--ovlp_ratio OVLP_RATIO
threshold of gene pair overlap regions identified by GMAP, default: 0.8
-b BLAST_COUNT, --blast_count BLAST_COUNT
blast count, default: 2
--blast_identity BLAST_IDENTITY
threshold of blast identity, default: 80
-e TE, --TE TE TE gff3 for filtering, default: ""
-j TE_OVERLAP, --TE_overlap TE_OVERLAP
threshold of TE overlap, default: 0.3, only effect when TE is not NULL
--paralog_only do TE filter only on paralog genes
-w WORKDIR, --workdir WORKDIR
workdir, default: wrkdir
-t THREADS, --threads THREADS
threads, default: 12Notice:
- the name of Chromosomes should be like: Chr01X, "X" means consecutive uppercase letters from A to Z, indicates different alleles, for example, if there are 4 alleles, the names should be: Chr01A,Chr01B,Chr01C,Chr01D.
- the gff3 files must contain "gene" records, or you can use "sed" command to change "mRNA" to "gene" for some downloaded gff3 files.
- there must no '-' in gene id.
-
Without TE filter
allele.adjusted.txt is the file contain all allele genes
allele.adjusted.*.stat are the statistics information of allele
-
With TE filter
allele.adjusted.nonTEs.txt is the file contain all allele genes
allele.adjusted.nonTEs.*.stat are the statistics information of allele
usage: allelefinder.py stat [-h] -i INPUT -g GFF3 -o OUTPUT
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
Input allele table
-g GFF3, --gff3 GFF3 GFF3 file of polyploid
-o OUTPUT, --output OUTPUT
Prefix of output file- Cleanup sequence
If user want to remove the duplicate genes with same CDS sequence, the "cleanup" would deal with them and only one gene would be kept randomly.
usage: allelefinder.py cleanup [-h] --in_cds IN_CDS [--in_pep IN_PEP] --in_gff3 IN_GFF3 --out_cds OUT_CDS [--out_pep OUT_PEP] --out_gff3 OUT_GFF3
options:
-h, --help show this help message and exit
--in_cds IN_CDS Input CDS file
--in_pep IN_PEP Input PEP file
--in_gff3 IN_GFF3 Input GFF3 file
--out_cds OUT_CDS Output CDS file
--out_pep OUT_PEP Output PEP file
--out_gff3 OUT_GFF3 Output GFF3 fileNotice: CDS file and GFF3 file are required, PEP file is optional.
- Adjust paralog genes
If there are too many genes be marked with paralog, you can use command below to pull them down as new alleles
usage: allelefinder.py adjust [-h] -i INPUT -m MIN_NUM -o OUTPUT
options:
-h, --help show this help message and exit
-i INPUT, --input INPUT
Input allele table
-m MIN_NUM, --min_num MIN_NUM
Minium number of genes, which means the number of genes marked as paralog that distribute in different allele should be pulled down as new allele genes
-o OUTPUT, --output OUTPUT
Output allele table