- Song WZ, Zhang S, Thomas T* (2022) MarkerMAG: linking metagenome-assembled genomes (MAGs) with 16S rRNA marker genes using paired-end short reads, Bioinformatics. https://doi.org/10.1093/bioinformatics/btac398
- Contact: Dr. Weizhi Song ([email protected]), Prof. Torsten Thomas ([email protected])
- Center for Marine Science & Innovation, University of New South Wales, Sydney, Australia
- 2022-05-08 - MarkerMAG is now available on Bioconda, please refers to "How to install" for details.
- 2022-03-12 - A demo dataset (together with command) has now been provided! You can use it to check if MarkerMAG is installed successfully on your system.
-
Main module
link: linking MAGs with 16S rRNA marker genes
-
Supplementary modules
-
MarkerMAG is implemented in python3, It has been tested on Linux and MacOS, but NOT on Windows.
-
A Conda package that automatically installs MarkerMAG's third-party dependencies (except Usearch
⚠️ ) is now available. Please note that you'll need to install Usearch on your own as it's not available in Conda due to license issue.# install with conda create -n MarkerMAG_env -c bioconda MarkerMAG # To activate the environment conda activate MarkerMAG_env # MarkerMAG is ready for running now, type "MarkerMAG -h" for help # To deactivate the environment conda deactivate -
It can also be installed with pip. Software dependencies need to be in your system path in this case. Dependencies for the
linkmodule include BLAST+, Barrnap, seqtk, Bowtie2, Samtools, HMMER, metaSPAdes and Usearch. Dependencies for the supplementary modules are provided in their corresponding manual page.# install with pip3 install MarkerMAG # upgrade with pip3 install --upgrade MarkerMAG -
Here are some example commands for UNSW Katana users.
-
⚠️ If you clone the repository directly off GitHub you might end up with a version that is still under development.
-
MarkerMAG’s input consists of
- A set of user-provided MAGs
- A set of 16S rRNA gene sequences (either user-provided or generated with the
matam_16smodule) - Input reads need to be quality-filtered and in fasta format (no quality score).
-
⚠️ MarkerMAG is designed to work with paired short-read data (i.e. Illumina). It assumes the id of reads in pair in the format ofXXXX.1andXXXX.2. The only difference is the last character. You can rename your reads with MarkerMAG'srename_readsmodule (manual). -
Although you can use your preferred tool to reconstruct 16S rRNA gene sequences from the metagenomic dataset, MarkerMAG does have a supplementary module (
matam_16s) to reconstruct 16S rRNA genes. Please refer to the manual here if you want to give it a go. -
Link 16S rRNA gene sequences with MAGs (demo dataset):
MarkerMAG link -p Demo -r1 demo_R1.fasta -r2 demo_R2.fasta -marker demo_16S.fasta -mag demo_MAGs -x fa -t 12
-
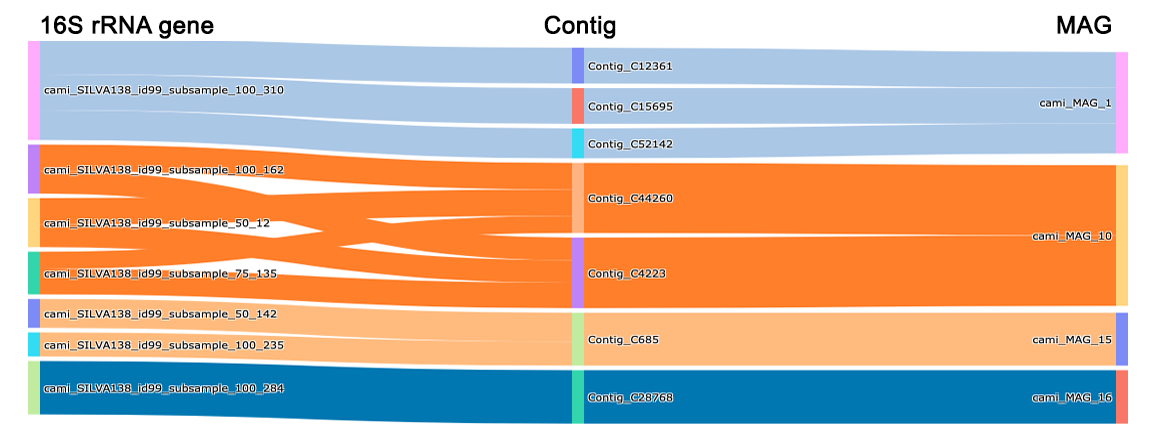
Summary of identified linkages at genome level:
Marker MAG Linkage Round matam_16S_7 MAG_6 181 Rd1 matam_16S_12 MAG_9 102 Rd1 matam_16S_6 MAG_59 55 Rd2 -
Summary of identified linkages at contig level (with figure):
Marker___MAG (linkages) Contig Round_1 Round_2 matam_16S_7___MAG_6(181) Contig_1799 176 0 matam_16S_7___MAG_6(181) Contig_1044 5 0 matam_16S_12___MAG_9(102) Contig_840 102 0 matam_16S_6___MAG_59(39) Contig_171 0 55 -
Copy number of linked 16S rRNA genes.
-
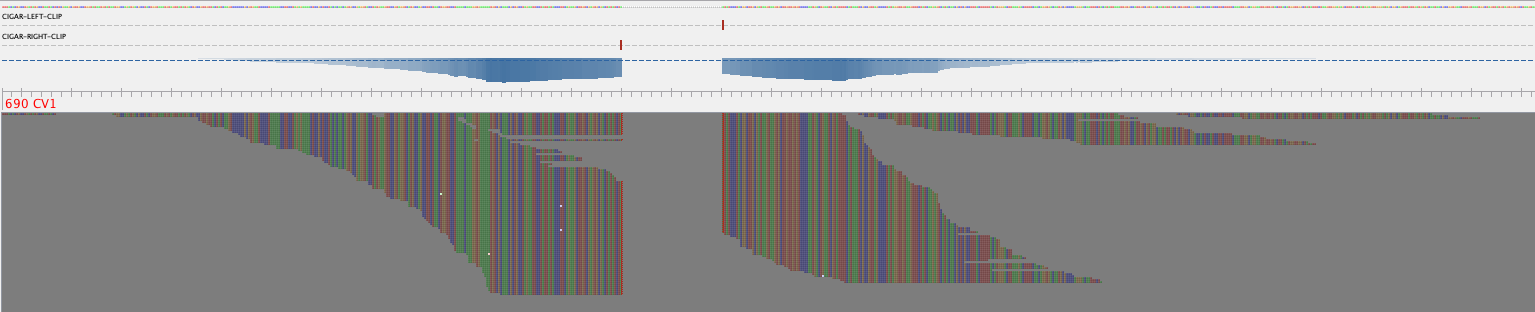
Visualization of individual linkage.
MarkerMAG supports the visualization of identified linkages (needs Tablet). Output files for visualization (example) can be found in the [Prefix]_linkage_visualization_rd1/2 folders. You can visualize how the linking reads are aligned to MAG contig and 16S rRNA gene by double-clicking the corresponding ".tablet" file. Fifty Ns are added between the linked MAG contig and 16S rRNA gene.
*If you saw error message from Tablet that says input files format can not be understood, please refer to here for a potential solution.