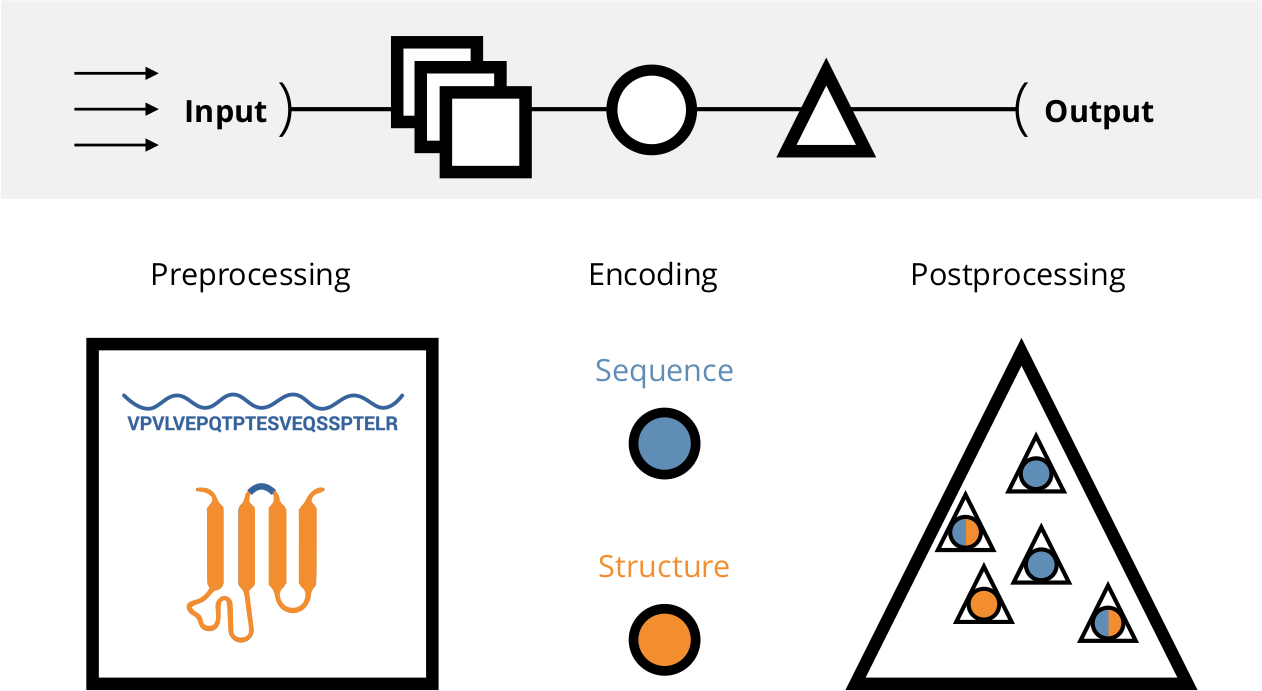
A workflow for an in-depth comparison and benchmarking of peptide encodings. All computations are highly parallelized and work efficiently across multiple datasets and encodings. For a thorough introduction refer to Spänig et al. (2021). The visualizations can be interactively accessed at https://peptidereactor.mathematik.uni-marburg.de/.
The emphasis is put on high-throughput processing of an arbitrary amount of input datasets (arrows), followed by the preprocessing, encoding, and postprocessing, generating the final output (top). The preprocessing includes sanitizing of the input sequences and the tertiary structure approximation, among others. Afterwards, the sequences as well as the accompanied structures are used for the encoding. The postprocessing involves machine learning and the visual preparation of the analyses, among others.
-
Clone this repo:
git clone [email protected]:spaenigs/peptidereactor.git. -
cdinto the root directory (peptidereactor/) -
Install conda:
peptidereactor/conda/install.sh. -
Create the conda environment:
peptidereactor/conda/create_env.sh -
Install docker:
- Ubuntu:
./peptidereactor/install_docker_io - Other distros:
./peptidereactor/install_docker_ce
- Ubuntu:
-
Build images:
./peptidereactor/docker/build_image./peptidereactor-vis/docker/build_image
-
Jobs to be executed:
./main.py --quiet --dag | dot -Tsvg > dag.svg(DAG)./main.py --quiet -nr(list)
-
Run the pipeline:
./main.py --quiet -
Results:
- Run server
./peptidereactor-vis/run_serverand - access http://localhost:8501
- Run server
The implementation of the PEPTIDE REACToR follows a modular design, such that meta nodes can be connected almost arbitrarily. Moreover, custom nodes can be easily added. Note that this tool is based on Snakemake, hence all conditions for a valid Snakemake-workflow must be also fulfilled.
A node to conduct multiple sequence alignment. Since we might need such a general node
multiple times, we add it to the utils category.
-
mkdir nodes/utils/multiple_sequence_alignment -
touch nodes/utils/multiple_sequence_alignment/Snakefile -
Specify input and output via config dictionary, e.g.,
config["fastas_in"]andconfig["fastas_out"]. -
Copy/paste into the
Snakefileand adapt stub:# from ... import ... TOKEN = config["token"] # access unique token rule all: input: config["fastas_out"] rule multiple_sequence_alignment: input: config["fastas_in"] output: config["fastas_out"] run: pass
-
touch nodes/utils/multiple_sequence_alignment/__init__.pyfor the API and copy/paste the following:import secrets # rule name def _get_header(token): return f''' rule utils_multiple_sequence_alignment_{token}:''' ... # specify input, output and path to the Snake- and configuration file. def _get_main(fastas_in, fastas_out): return f''' input: fastas_in={fastas_in} output: fastas_out={fastas_out} ... ''' # specify input and output def rule(fastas_in, fastas_out, benchmark_dir=None): token = secrets.token_hex(4) rule = _get_header(token) if benchmark_dir is not None: benchmark_out = f"{benchmark_dir}utils_multiple_sequence_alignment_{token}.txt" ...
Refer to an actual
__init__.pyfor a complete example. -
Make the node visible by adding
from . import multiple_sequence_alignmentinnodes/utils/__init__.py. -
Import and use the node in
main.py:import nodes.utils as utils w.add(utils.multiple_sequence_alignment.rule( fastas_in=["data/{dataset}/seqs_mapped.fasta", "data/{dataset}/seqs_sec.fasta", "data/{dataset}/seqs_ter.fasta"], fastas_out=["data/{dataset}/seqs_msa.fasta", "data/{dataset}/seqs_msa_sec.fasta", "data/{dataset}/seqs_msa_ter.fasta"], benchmark_dir=w.benchmark_dir))
-
During implementation, it might be helpful to run the rule isolated:
./peptidereactor/run_pipeline -s nodes/utils/multiple_sequence_alignment/Snakefile \ --config fasta_in=... fasta_out=... token=... \ -nror even access the Docker container interactively:
docker run -it --entrypoint "/bin/bash" peptidereactor
Note that the PEPTIDE REACToR is still under development, thus changes are likely. However, the fundamental structure, highlighted above, will remain.