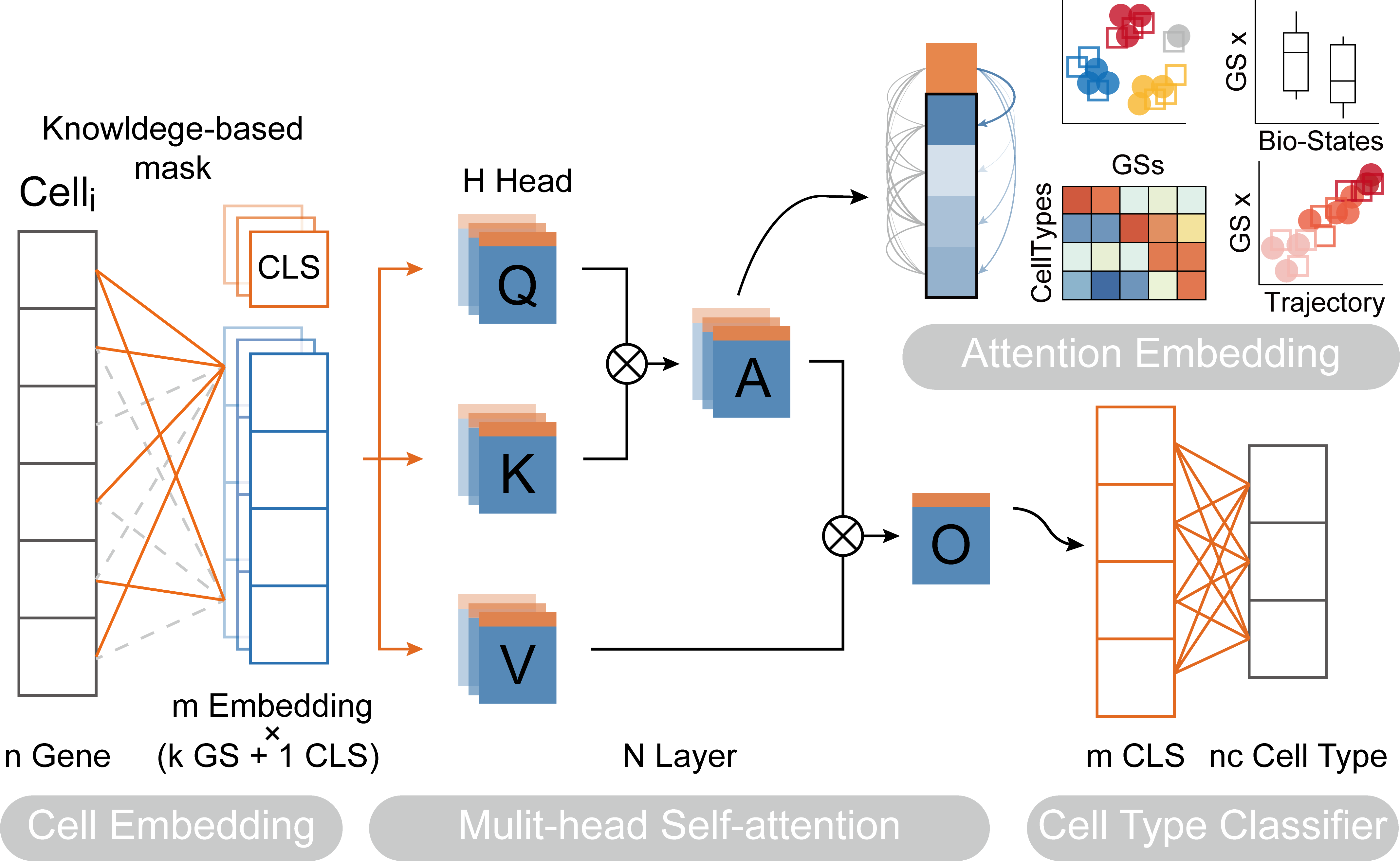
We created the python package called TOSICA that uses scanpy ans torch to explainablely annotate cell type on single-cell RNA-seq data.
- Linux/UNIX/Windows system
- Python >= 3.8
- torch == 1.7.1
conda create -n TOSICA python=3.8 scanpy
conda activate TOSICA
conda install pytorch=1.7.1 torchvision=0.8.2 torchaudio=0.7.2 cudatoolkit=10.1 -c pytorch
The TOSICA python package is in the folder TOSICA. You can simply install it from the root of this repository using
pip install .
Alternatively, you can also install the package directly from GitHub via
pip install git+https://github.com/JackieHanLab/TOSICA.git
TOSICA.yaml
TOSICA.train(ref_adata, gmt_path,project=<my_project>,label_name=<label_key>)ref_adata: anAnnDataobject of reference dataset.gmt_path: default pre-prepared mask or path to .gmt files.<my_project>: the model will be saved in a folder named <my_project>. Default:<gmt_path>_20xxxxxx.<label_key>: the name of the label column inref_adata.obs.
human_gobp: GO_bp.gmthuman_immune: immune.gmthuman_reactome: reactome.gmthuman_tf: TF.gmtmouse_gobp: m_GO_bp.gmtmouse_reactome: m_reactome.gmtmouse_tf: m_TF.gmt
./my_project/mask.npy: Mask matrix./my_project/pathway.csv: Gene set list./my_project/label_dictionary.csv: Label list./my_project/model-n.pth: Weights
new_adata = TOSICA.pre(query_adata, model_weight_path = <path to optional weight>,project=<my_project>)query_adata: anAnnDataobject of query dataset .model_weight_path: the weights generated duringscTrans.train, like:'./weights20220607/model-6.pth'.project: name of the folder build in training step, like:my_projector<gmt_path>_20xxxxxx.
new_adata.X: Attention matrixnew_adata.obs['Prediction']: Predicted labelsnew_adata.obs['Probability']: Probability of the predictionnew_adata.var['pathway_index']: Gene set of each colume./my_project/gene2token_weights.csv: The weights matrix of genes to tokens
Warning: the
var_names(genes) of theref_adataandquery_adatamust be consistent and in the same order.query_adata = query_adata[:,ref_adata.var_names]Please run the code to make sure they are the same.