CAISC, which stands for Clonal Architecture with Integration of SNV and CNV, is a software that integrates copy number variation and single nucleotide mutations for genetic heterogeneity profiling and subclone detection by single-cell RNA sequencing
If you have any questions about using CAISC, you can open a new issue here, or you can email one of the maintainers listed below.
Install to R/RStudio and install all packages in the latest version of R.
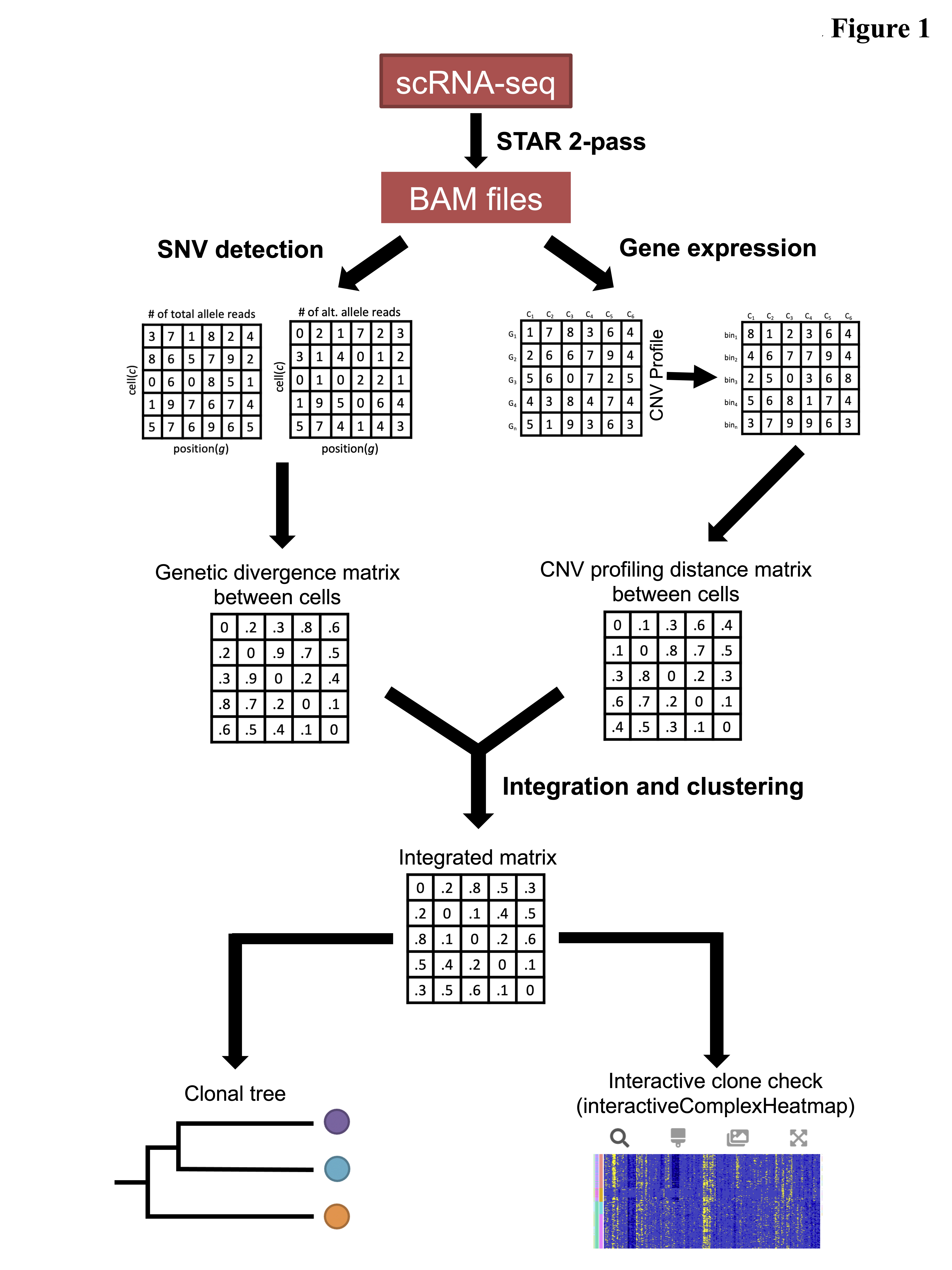
devtools::install_github("lizamathews/CAISC")We present Clonal Architecture with Integration of SNV and CNV (CAISC), a new subclone detection algorithm that integrates both SNV and CNV data in order to acquire more accurate and robust clustering results. Here is an overview of the CAISC pipeline. We derive two cell-cell distance matrices using either SNV and CNV data, from DENDRO and infercnv respectively. These matrices are integrated using an entropy weighted method, and into a final distance matrix that is used to cluster the cells into subclones.
Figure 1. Clonal Architecture with Integration of SNV and CNV (CAISC). Overview of the computational framework that integrates both SNV and CNV profiles for clonal identification from scRNA-seq data. scRNA-seq reads are aligned with STAR, mutations are identified with GATK, and the read counts were calculated to represent gene expression. DENDRO is used to calculate a distance matrix based on allele reads of mutations. Infercnv is used to convert gene expression level data to a CNV profile matrix and then to calculate a distance matrix between cells. These two distance matrices are integrated using an entropy weighted method. Finally, a clonal tree is generated using the integrated matrix to more accurately identify subclones and infer their evolutionary relationship. The interactive figure generated by interactiveComplexHeatmap allows for manual examination of inferred clones.
The CAISC R notebook with step-by-step sample code is available here.
The corresponding Rmd script is available here.
CAISC: A Software to Integrate Copy Number Variation and Single Nucleotide Mutations for Genetic Heterogeneity Profiling and Subclone Detection by Single-cell RNA Sequencing (GitHub: https://github.com/lizamathews/CAISC)
- Shouguo Gao ([email protected])
- Jeerthi Kannan ([email protected])
- Liza Mathews ([email protected])