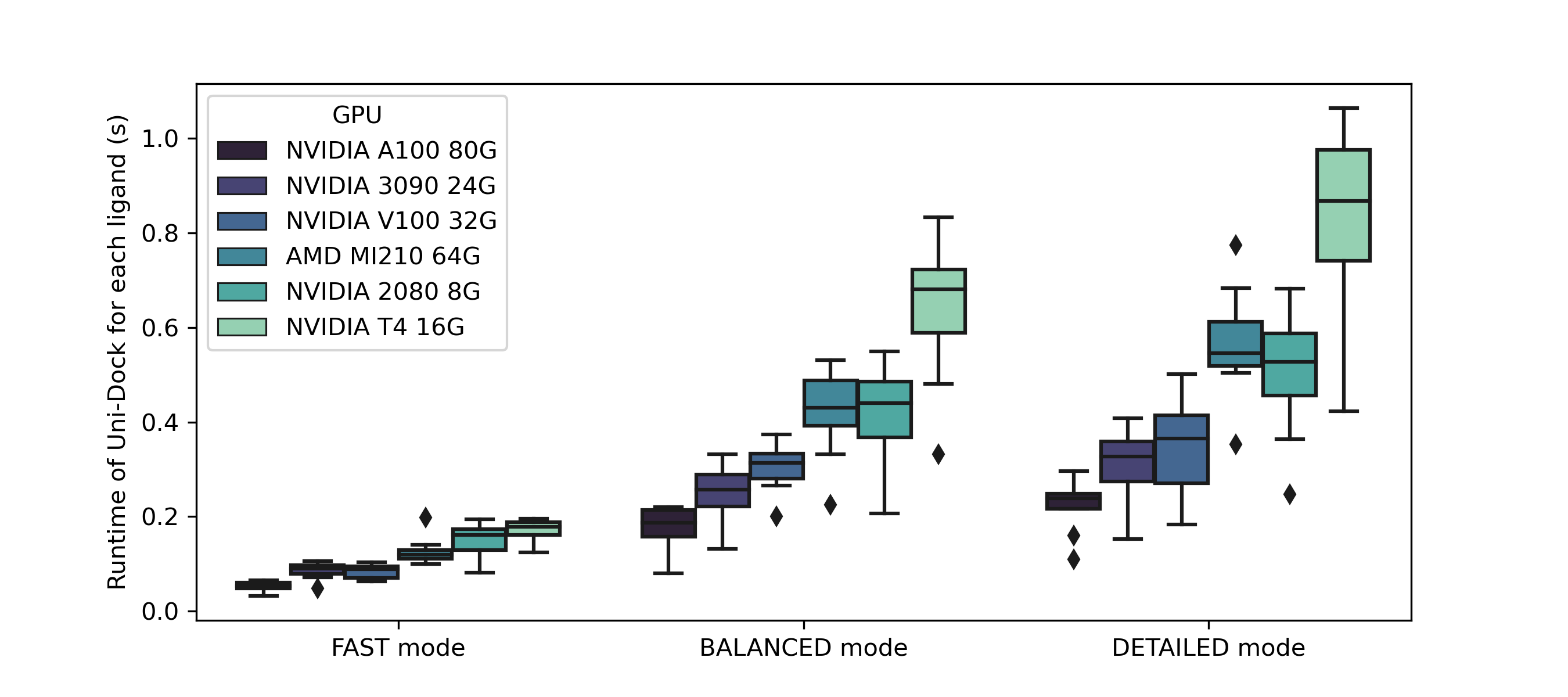
Uni-Dock is a GPU-accelerated molecular docking program developed by DP Technology. It supports various scoring functions including vina, vinardo, and ad4. Uni-Dock achieves more than 1000-fold speed-up on V100 GPU with high-accuracy compared with the AutoDock Vina running in single CPU core. The paper has been accepted by JCTC (doi: 10.1021/acs.jctc.2c01145).
- V1.1.0: Support SDF format input for vina and vinardo scoring functions.
We offer the software for academic purposes only. By downloading and using Uni-Dock, you are agreeing to the usage guideline (en, zh).
Developed by DP Technology, Hermite® is a new-generation drug computing design platform which integrates artificial intelligence, physical modeling and high-performance computing to provide a one-stop computing solution for preclinical drug research and development. It integrates the features of Uni-Dock, along with virtual screening workflow for an efficient drug discovery process.
Uni-Dock is now available on the new-generation drug computing design platform Hermite® for ultralarge virtual screening.
For commercial usage and further cooperations, please contact us at [email protected] .
Uni-Dock supports NVIDIA GPUs on Linux platform. CUDA toolkit is required.
Please download the latest binary of Uni-Dock at the assets tab of the Release page.
Executable unidock supports vina and vinardo scoring functions, and unidock_ad4 supports ad4 scoring function.
After downloading, please make sure that the path to unidock is in your PATH environment variable.
To launch a Uni-Dock job, the most important parameters are as follows:
--receptor: filepath of the receptor (PDBQT)--gpu_batch: filepath of the ligands to dock with GPU (PDBQT), enter multiple at a time, separated by spaces (" ")--search_mode: computational complexity, choice in [fast,balance, anddetail].
Advanced options
--search_mode is the recommended setting of --exhaustiveness and --max_step, with three combinations called fast, balance, and detail.
fastmode:--exhaustiveness 128&--max_step 20balancemode:--exhaustiveness 384&--max_step 40detailmode:--exhaustiveness 512&--max_step 40
The larger --exhaustiveness and --max_step, the higher the computational complexity, the higher the accuracy, but the larger the computational cost.
unidock --receptor <receptor.pdbqt> \
--gpu_batch <lig1.pdbqt> <lig2.pdbqt> ... <ligN.pdbqt> \
--search_mode balance \
--scoring vina \
--center_x <center_x> \
--center_y <center_y> \
--center_z <center_z> \
--size_x <size_x> \
--size_y <size_y> \
--size_z <size_z> \
--num_modes 1 \
--dir <save dir>>> unidock --help
Input:
--receptor arg rigid part of the receptor (PDBQT or PDB)
--flex arg flexible side chains, if any (PDBQT or PDB)
--ligand arg ligand (PDBQT)
--ligand_index arg file containing paths to ligands (PDBQT or SDF)
--batch arg batch ligand (PDBQT)
--gpu_batch arg gpu batch ligand (PDBQT or SDF)
--scoring arg (=vina) scoring function (ad4, vina or vinardo)
Search space (required):
--maps arg affinity maps for the autodock4.2 (ad4) or vina
scoring function
--center_x arg X coordinate of the center (Angstrom)
--center_y arg Y coordinate of the center (Angstrom)
--center_z arg Z coordinate of the center (Angstrom)
--size_x arg size in the X dimension (Angstrom)
--size_y arg size in the Y dimension (Angstrom)
--size_z arg size in the Z dimension (Angstrom)
--autobox set maps dimensions based on input ligand(s) (for
--score_only and --local_only)
Output (optional):
--out arg output models (PDBQT), the default is chosen based
on the ligand file name
--dir arg output directory for batch mode
--write_maps arg output filename (directory + prefix name) for
maps. Option --force_even_voxels may be needed to
comply with .map format
Misc (optional):
--cpu arg (=0) the number of CPUs to use (the default is to try
to detect the number of CPUs or, failing that, use
1)
--seed arg (=0) explicit random seed
--exhaustiveness arg (=8) exhaustiveness of the global search (roughly
proportional to time): 1+
--max_evals arg (=0) number of evaluations in each MC run (if zero,
which is the default, the number of MC steps is
based on heuristics)
--num_modes arg (=9) maximum number of binding modes to generate
--min_rmsd arg (=1) minimum RMSD between output poses
--energy_range arg (=3) maximum energy difference between the best binding
mode and the worst one displayed (kcal/mol)
--spacing arg (=0.375) grid spacing (Angstrom)
--verbosity arg (=1) verbosity (0=no output, 1=normal, 2=verbose)
--max_step arg (=0) maximum number of steps in each MC run (if zero,
which is the default, the number of MC steps is
based on heuristics)
--refine_step arg (=5) number of steps in refinement, default=5
--max_gpu_memory arg (=0) maximum gpu memory to use (default=0, use all
available GPU memory to optain maximum batch size)
--search_mode arg search mode of unidock (fast, balance, detail), using
recommended settings of exhaustiveness and search
steps; the higher the computational complexity,
the higher the accuracy, but the larger the
computational cost
Configuration file (optional):
--config arg the above options can be put here
Information (optional):
--help display usage summary
--help_advanced display usage summary with advanced options
--version display program versionWe have provided a target from DUD-E dataset for screening test. Python version >=3.6 is recommended.
git clone https://github.com/dptech-corp/Uni-Dock.git
cd Uni-Dock/example/screening_test
# target def
cp config_def.json config.json
python run_dock.py
# target mmp13
cp config_mmp13.json config.json
python run_dock.pyIf you want to use search mode presets, specify the parameter search_mode in config.json and delete nt and ns in config.json.
Please report bugs to Issues page.
If you used Uni-Dock in your work, please cite:
Yu, Y., Cai, C., Wang, J., Bo, Z., Zhu, Z., & Zheng, H. (2023). Uni-Dock: GPU-Accelerated Docking Enables Ultralarge Virtual Screening. Journal of Chemical Theory and Computation. https://doi.org/10.1021/acs.jctc.2c01145
Tang, S., Chen, R., Lin, M., Lin, Q., Zhu, Y., Ding, J., ... & Wu, J. (2022). Accelerating autodock vina with gpus. Molecules, 27(9), 3041. DOI 10.3390/molecules27093041
J. Eberhardt, D. Santos-Martins, A. F. Tillack, and S. Forli AutoDock Vina 1.2.0: New Docking Methods, Expanded Force Field, and Python Bindings, J. Chem. Inf. Model. (2021) DOI 10.1021/acs.jcim.1c00203
O. Trott, A. J. Olson, AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading, J. Comp. Chem. (2010) DOI 10.1002/jcc.21334
- The GPU encounters out-of-memory error.
Uni-Dock estimates the number of ligands put into GPU memory in one pass based on the available GPU memory size. If it fails, please use
--max_gpu_memoryto limit the usage of GPU memory size by Uni-Dock. - I want to put all my ligands in
--gpu_batch, but it exceeds the maximum command line length that linux can accept.- You can save your command in a shell script like
run.sh, and run the command bybash run.sh. - You can save your ligands path in a file (separated by spaces) by
ls *.pdbqt > index.txt, and use--ligand_index index.txtin place of--gpu_batch.
- You can save your command in a shell script like