Main features • Installation • Overview • Databases • Example workflow • Analysis across trials • Tests • Acknowledgements • Future
The package ctrdata provides functions for retrieving (downloading)
information on clinical trials from public registers, and for
aggregating and analysing this information; it can be used for the
- EU Clinical Trials Register (“EUCTR”, https://www.clinicaltrialsregister.eu/)
- ClinicalTrials.gov (“CTGOV” or 🔔“CTGOV2”, see example)
- ISRCTN (https://www.isrctn.com/)
- EU Clinical Trials Information System (“CTIS”, https://euclinicaltrials.eu/) 🔔 see example
The motivation is to investigate and understand trends in design and
conduct of trials, their availability for patients and to facilitate
using their detailed results for research and meta-analyses. ctrdata
is a package for the R system, but other
systems and tools can be used with the databases created with the
package. This README was reviewed on 2023-09-17 for version 1.15.2.9000
for CTGOV changes.
- Protocol- and results-related trial information is easily downloaded:
Users define a query in a register’s web interface and then enter the
URL into
ctrdatawhich retrieves in one go all trials found. Documents in registers on trials can also be downloaded. Personal annotations can be made to trials when downloading a query. Synonyms of an active substance can also be found. - Downloaded trial information is transformed and stored in a collection
of a document-centric database, for fast and offline access.
Information from different registers can be accumalated in a single
collection. Uses
DuckDB,PostgreSQL,RSQLiteorMongoDB, via R packagenodbi: see section Databases below. Re-run any previous query in a collection to retrieve and update trial records. - For analyses, convenience functions in
ctrdataallow to identify unique (de-duplicated) trial records across registers, to merge and recode fields as well as to easily access deeply-nested fields. Analysis can be done withRor other systems, by accessing the structured information in the database.
URLs of queries in the registers can be automatically copied to the clipboard (including for “CTIS”, where the URL does not show the query), see here.
Remember to respect the registers’ terms and conditions (see
ctrOpenSearchPagesInBrowser(copyright = TRUE)). Please cite this
package in any publication as follows: “Ralf Herold (2023). ctrdata:
Retrieve and Analyze Clinical Trials in Public Registers. R package
version 1.14.0, https://cran.r-project.org/package=ctrdata”.
Package ctrdata has been used for:
- Lasch et al. (2022) The Impact of COVID‐19 on the Initiation of Clinical Trials in Europe and the United States. Clinical Pharmacology & Therapeutics, https://doi.org/10.1002/cpt.2534
- Blogging on Innovation coming to paediatric research
- Cancer Research UK (2017) The impact of collaboration: The value of UK medical research to EU science and health
Package ctrdata is on
CRAN and on
GitHub. Within
R, use the following commands to install
package ctrdata:
# Install CRAN version:
install.packages("ctrdata")
# Alternatively, install development version:
install.packages("devtools")
devtools::install_github("rfhb/ctrdata", build_vignettes = TRUE)These commands also install the package’s dependencies (nodbi,
jsonlite, httr, curl, clipr, xml2, rvest, lubridate and
stringi).
These are required for ctrLoadQueryIntoDb(), the main function of
package ctrdata (see Example workflow), to work
with the registers EUCTR, CTGOV, ISRCTN (but are not required to work
with CTIS); the function also checks if the tools can be used.
-
For MS Windows, install
Cygwin: InR, runctrdata::installCygwinWindowsDoInstall()for an automated minimal installation. Alternatively, manually install Cygwin with packagesperl,php-jsoncandphp-simplexmlintoc:\cygwin. The installation needs about 160 MB disk space. -
In macOS including 11 Big Sur, these are already installed; as alternative and 🔔for macOS 12 Monterey and above,
homebrewcan be used:brew install php, which seems to include allphplibraries required forctrdata. -
In Linux, these are usually already installed; tools to install vary by distribution (e.g.,
sudo apt install php-cli php-xml php-json).
This is optional; it works with all registers supported by ctrdata but
is recommended for CTIS because the URL in the web browser does not
reflect the parameters the user specified for querying this register.
In the web browser, install the Tampermonkey browser
extension, click on “New user script”
and then on “Tools”, then enter into “Import from URL” this URL:
https://raw.githubusercontent.com/rfhb/ctrdata/master/tools/ctrdataURLcopier.js
and last click on “Install”.
The browser extension can be disabled and enabled by the user. When
enabled, the URLs to all user’s queries in the registers are
automatically copied to the clipboard and can be pasted into the
queryterm=... parameter of function ctrLoadQueryIntoDb().
The functions are listed in the approximate order of use in a user’s workflow (in bold, main functions). See also the package documentation overview.
| Function name | Function purpose |
|---|---|
ctrOpenSearchPagesInBrowser() |
Open search pages of registers or execute search in web browser |
ctrFindActiveSubstanceSynonyms() |
Find synonyms and alternative names for an active substance |
ctrGetQueryUrl() |
Import from clipboard the URL of a search in one of the registers |
ctrLoadQueryIntoDb() |
Retrieve (download) or update, and annotate, information on trials from a register and store in a collection in a database |
dbQueryHistory() |
Show the history of queries that were downloaded into the collection |
dbFindIdsUniqueTrials() |
Get the identifiers of de-duplicated trials in the collection |
dbFindFields() |
Find names of variables (fields) in the collection |
dbGetFieldsIntoDf() |
Create a data frame (or tibble) from trial records in the database with the specified fields |
dfTrials2Long() |
Transform the data.frame from dbGetFieldsIntoDf() into a long name-value data.frame, including deeply nested fields |
dfName2Value() |
From a long name-value data.frame, extract values for variables (fields) of interest (e.g., endpoints) |
dfMergeVariablesRelevel() |
Merge variables into a new variable, optionally map values to a new set of levels |
installCygwinWindowsDoInstall() |
Convenience function to install a Cygwin environment (MS Windows only) |
Package ctrdata retrieves trial information and stores it in a
database collection, which has to be given as a connection object to
parameter con for several ctrdata functions; this connection object
is created in slightly different ways for the four supported database
backends that can be used with ctrdata as shown in the table. For a
speed comparison, see the nodbi
documentation.
Besides ctrdata functions below, any such a connection object can
equally be used with functions of other packages, such as nodbi (last
row in table) or, in case of MongoDB as database backend, mongolite
(see vignettes).
| Purpose | Function call |
|---|---|
| Create SQLite database connection | dbc <- nodbi::src_sqlite(dbname = "name_of_my_database", collection = "name_of_my_collection") |
| Create MongoDB database connection | dbc <- nodbi::src_mongo(db = "name_of_my_database", collection = "name_of_my_collection") |
| Create PostgreSQL database connection | dbc <- nodbi::src_postgres(dbname = "name_of_my_database"); dbc[["collection"]] <- "name_of_my_collection" |
| Create DuckDB database connection | dbc <- nodbi::src_duckdb(dbdir = "name_of_my_database", collection = "name_of_my_collection") |
Use connection with ctrdata functions |
ctrdata::{ctrLoadQueryIntoDb, dbQueryHistory, dbFindIdsUniqueTrials, dbFindFields, dbGetFieldsIntoDf}(con = dbc, ...) |
Use connection with nodbi functions |
e.g., nodbi::docdb_query(src = dbc, key = dbc$collection, ...) |
- Install R package ctrdata
- Retrieve clinical trial information
- Summarise and analyse clinical trial information
The aim is to download protocol-related trial information and tabulate the trials’ status of conduct.
- Attach package
ctrdata:
library(ctrdata)- See help to get started with
ctrdata:
help("ctrdata")- Information on trial registers that can be used with
ctrdata:
help("ctrdata-registers")- Open registers’ advanced search pages in browser:
ctrOpenSearchPagesInBrowser()
# Please review and respect register copyrights:
ctrOpenSearchPagesInBrowser(copyright = TRUE)-
Adjust search parameters and execute search in browser
-
When trials of interest are listed in browser, copy the address from the browser’s address bar to the clipboard
-
Search used in this example: https://www.clinicaltrialsregister.eu/ctr-search/search?query=cancer&age=under-18&phase=phase-one&status=completed
-
Get address from clipboard:
q <- ctrGetQueryUrl()
# * Using clipboard content as register query URL: https://www.clinicaltrialsregister.eu/ctr-search/search?query=cancer&age=under-18&phase=phase-one&status=completed
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one&status=completed
q
# query-term query-register
# 1 query=cancer&age=under-18&phase=phase-one&status=completed EUCTR🔔 Queries in the trial registers can automatically copied to the clipboard (including for “CTIS”, where the URL does not show the query) using our solution here.
- Retrieve protocol-related information, transform and save to database:
The database collection is specified first, using nodbi (see above for
how to specify PostgreSQL, RSQlite, DuckDB or MongoDB as
backend, see section
Databases); then, trial
information is retrieved and loaded into the collection:
# Connect to (or newly create) an SQLite database
# that is stored in a file on the local system:
db <- nodbi::src_sqlite(
dbname = "some_database_name.sqlite_file",
collection = "some_collection_name"
)
# Retrieve trials from public register:
ctrLoadQueryIntoDb(
queryterm = q,
euctrresults = TRUE,
con = db
)
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one&status=completed
# Checking helper binaries: . . . . . done
# * Checking trials in EUCTR...
# Retrieved overview, multiple records of 92 trial(s) from 5 page(s) to be downloaded (estimate: 4.6 MB)
# (1/3) Downloading trials...
# Note: register server cannot compress data, transfer takes longer, about 0.2s per trial
# Download status: 5 done; 0 in progress. Total size: 7.33 Mb (100%)... done!
# (2/3) Converting to JSON, 353 records converted
# (3/3) Importing JSON records into database...
# = Imported or updated 353 records on 92 trial(s)
# * Checking results if available from EUCTR for 92 trials:
# (1/4) Downloading and extracting results (. = data, F = file[s] and data, x = none):
# Download status: 92 done; 0 in progress. Total size: 57.94 Mb (100%)... done!
# F . F F . F . F . . . . F F . . . . . . F . . . . . . . . . . . . . . . . . . . . . . . . . . . .
# . . F . . . . F . . F . . . . . . F . . . . F . , 74 records converted
# (3/4) Importing JSON into database...
# (4/4) Results history: not retrieved (euctrresultshistory = FALSE)
# = Imported or updated results for 74 trials
# No history found in expected format.
# Updated history ("meta-info" in "some_collection_name")Under the hood, scripts euctr2json.sh and xml2json.php (in
ctrdata/exec) transform EUCTR plain text files and CTGOV as well as
ISRCTN XML files to ndjson format, which is imported into the
database collection.
- Analyse
Tabulate the status of trials that are part of an agreed paediatric
development program (paediatric investigation plan, PIP). ctrdata
functions return a data.frame (or a tibble, if package tibble is
loaded).
# Get all records that have values in the fields of interest:
result <- dbGetFieldsIntoDf(
fields = c(
"a7_trial_is_part_of_a_paediatric_investigation_plan",
"p_end_of_trial_status",
"a2_eudract_number"
),
con = db
)
# Find unique (deduplicated) trial identifiers for trials that have more than
# one record, for example for several EU Member States or in several registers:
uniqueids <- dbFindIdsUniqueTrials(con = db)
# Searching for duplicate trials...
# - Getting all trial identifiers (may take some time), 353 found in collection
# - Finding duplicates among registers' and sponsor ids...
# - 261 EUCTR _id were not preferred EU Member State record for 92 trials
# - Keeping 92 / 0 / 0 / 0 / 0 records from EUCTR / CTGOV / CTGOV2 / ISRCTN / CTIS
# = Returning keys (_id) of 92 records in collection "some_collection_name"
# Keep only unique / de-duplicated records:
result <- subset(
result,
subset = `_id` %in% uniqueids
)
# Tabulate the selected clinical trial information:
with(
result,
table(
p_end_of_trial_status,
a7_trial_is_part_of_a_paediatric_investigation_plan
)
)
# a7_trial_is_part_of_a_paediatric_investigation_plan
# p_end_of_trial_status FALSE TRUE
# Completed 46 20
# GB - no longer in EU/EEA 1 1
# Ongoing 3 1
# Prematurely Ended 3 2
# Restarted 0 1
# Temporarily Halted 1 1- Add records from another register (CTGOV) into the same collection
🔔Both the current and classic CTGOV website are supported as of
2023-08-05. The new website and API introduced in July 2023
(https://www.clinicaltrials.gov/) is identified in ctrdata as
CTGOV2. The website and API which is now called “classic”
(https://classic.clinicaltrials.gov/) is identified in ctrdata as
CTGOV, and this is backwards-compatible with queries that were
previously retrieved with ctrdata.
Both use the same trial identifier (e.g., NCT01234567) for the same
trial. As a consequence, queries for the same trial retrieved using
CTGOV or CTGOV2 overwrite any previous record for that trial,
whether loaded from CTGOV or CTGOV2. Thus, only a single version
(the last retrieved) will be in the collection in the user’s database.
Important differences exist between field names and contents of
information retrieved using CTGOV or CTGOV2; see the XML schemas
for CTGOV
and the REST API for
CTGOV2.
For more details, call help("ctrdata-registers"). This is one of the
reasons why ctrdata handles the situation as if these were two
different registers.
- Search used in this example: https://www.clinicaltrials.gov/search?cond=Neuroblastoma&aggFilters=ages:child,results:with,studyType:int
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = "cond=Neuroblastoma&aggFilters=ages:child,results:with,studyType:int",
register = "CTGOV2",
con = db
)
# * Appears specific for CTGOV REST API 2.0.0
# * Found search query from CTGOV2: cond=Neuroblastoma&aggFilters=ages:child,results:with,studyType:int
# * Checking trials using CTGOV API 2.0.0.-test, found 88 trials
# (1/2) Downloading and converting in 1 batch(es) (max. 1000 trials each; estimate: 8.8 MB total)
# Download status: 1 done; 0 in progress. Total size: 8.75 Mb (837%)...converting to NDJSON...
# (2/2) Importing JSON records into database...
# JSON file #: 1 / 1
# = Imported / updated 88 records on 88 trial(s)
# Updated history ("meta-info" in "some_collection_name")- Using an example from classic CTGOV: https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = "https://classic.clinicaltrials.gov/ct2/results?cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug",
con = db
)
# * Appears specific for CTGOV CLASSIC
# * Found search query from CTGOV: cond=neuroblastoma&rslt=With&recrs=e&age=0&intr=Drug
# Checking helper binaries: done
# * Checking trials in CTGOV classic...
# Retrieved overview, records of 55 trial(s) are to be downloaded (estimate: 0.44 MB)
# (1/3) Downloading trial file...
# Download status: 1 done; 0 in progress. Total size: 755.00 Kb (100%)... done!
# (2/3) Converting to JSON, 55 records converted
# (3/3) Importing JSON records into database...
# = Imported or updated 55 trial(s)
# Updated history ("meta-info" in "some_collection_name")- Add records from a third register (ISRCTN) into the same collection
Search used in this example: https://www.isrctn.com/search?q=neuroblastoma
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = "https://www.isrctn.com/search?q=neuroblastoma",
con = db
)
# * Found search query from ISRCTN: q=neuroblastoma
# Checking helper binaries: done
# * Checking trials in ISRCTN...
# Retrieved overview, records of 9 trial(s) are to be downloaded (estimate: 0.16 MB)
# (1/3) Downloading trial file...
# Download status: 1 done; 0 in progress. Total size: 93.10 Kb (100%)... done!
# (2/3) Converting to JSON, 9 records converted
# (3/3) Importing JSON records into database...
# = Imported or updated 9 trial(s)
# Updated history ("meta-info" in "some_collection_name")- Add records from a fourth register (CTIS 🔔) into the same collection
Queries in the CTIS search interface can be automatically copied to the
clipboard so that a user can paste them into queryterm, see
here. As
of June 2023, more than 200 trials are publicly accessible in CTIS. See
below for how to download documents from CTIS.
# Retrieve trials from another register:
ctrLoadQueryIntoDb(
queryterm = "https://euclinicaltrials.eu/app/#/search?ageGroupCode=2",
con = db
)
# * Found search query from CTIS: ageGroupCode=2
# * Checking trials in EUCTR...
# (1/5) Downloading trials list, found 32 trials
# (2/5) Downloading and processing part I and parts II... (estimate: 4.8 Mb)
# Download status: 32 done; 0 in progress. Total size: 4.94 Mb (100%)... done!
# . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . . .
# (3/5) Downloading and processing additional data:
# publicevents, summary, layperson, csr, cm, inspections, publicevaluation
# Download status: 46 done; 0 in progress. Total size: 1.84 Mb (100%)... done!
# 32
# (4/5) Importing JSON records into database...
# (5/5) Updating with additional data: . .
# = Imported / updated 32 / 32 / 32 records on 32 trial(s)
# Updated history ("meta-info" in "some_collection_name")
allFields <- dbFindFields(".*", db)
# Finding fields in database collection (may take some time)
# Field names cached for this session.
length(allFields[grepl("CTIS", names(allFields))])
# [1] 1886
allFields[grepl("defer|consideration$", allFields, ignore.case = TRUE)]
# CTIS
# "hasDeferrallApplied"
# CTIS
# "publicEvaluation.partIIEvaluationList.partIIRfiConsiderations.rfiConsiderations.consideration"
# CTIS
# "publicEvaluation.partIRfiConsiderations.rfiConsiderations.consideration"
# CTIS
# "publicEvaluation.partIRfiConsiderations.rfiConsiderations.part1Consideration"
# CTIS
# "publicEvaluation.validationRfiConsiderations.rfiConsiderations.consideration"
dbGetFieldsIntoDf("publicEvaluation.partIRfiConsiderations.rfiConsiderations.consideration", db)[1,2]
# publicEvaluation.partIRfiConsiderations.rfiConsiderations.consideration
# In(EX)clusion criteria: An adequate definition of WOCBP or postmenopausal woman
# is missing and should be added to the protocol. / The rationale for the treatment duration of 7 to
# 18 weeks cannot be followed. No data are available for this short time period and nivolumab treatment.
# The shortest duration tested so far in 1 year in adjuvant or maintenance treatment protocol.
# The sponsor is asked to justify and substantiate his assumption that this treatment duration is
# adequate with respective data. / E: Information regarding the special clinical conditions for
# conducting clinical trials with minors, \nsee Article 32 Par. 1 lit e) to g) of Regulation (EU)
# 536/2014 is missing. Please revise the protocol accordingly. So it is indicated to include the
# patients older than 18 years first and in case of positive results the planned younger patients
# could follow.\nStatistical Comment: The statistical analyses are missing in the trial protocol.
# Biometric adequate is a restriction to descriptive evaluations. A sequential evaluation is
# recommended (adults first and then children). The protocol has to be amended accordingly /
# Discontinuation criteria for study subjects and clinical trial termination criteria are missing
# and have to be added. Please amend. [...]
# use an alternative to dbGetFieldsIntoDf()
allData <- nodbi::docdb_query(src = db, key = db$collection, query = '{"ctrname":"CTIS"}')
# names of top-level data items
sort(names(allData))
# [1] "_id" "ageGroup" "applications"
# [4] "authorizationDate" "authorizedPartI" "authorizedPartsII"
# [7] "coSponsors" "ctNumber" "ctrname"
# [10] "ctStatus" "decisionDate" "eeaStartDate"
# [13] "eudraCtInfo" "gender" "hasAmendmentApplied"
# [16] "hasDeferrallApplied" "id" "initialApplicationId"
# [19] "isRmsTacitAssignment" "lastUpdated" "memberStatesConcerned"
# [22] "mscTrialNotificationsInfoList" "primarySponsor" "publicEvaluation"
# [25] "record_last_import" "recruitmentStatus" "sponsorType"
# [28] "startDateEU" "submissionDate" "therapeuticAreas"
# [31] "title" "totalNumberEnrolled" "totalPartIISubjectCount"
# [34] "trialCountries" "trialGlobalEnd" "trialPhase"
# [37] "trialStartDate"
#
format(object.size(allData), "MB")
# [1] "25.5 Mb"- Analysis across trials
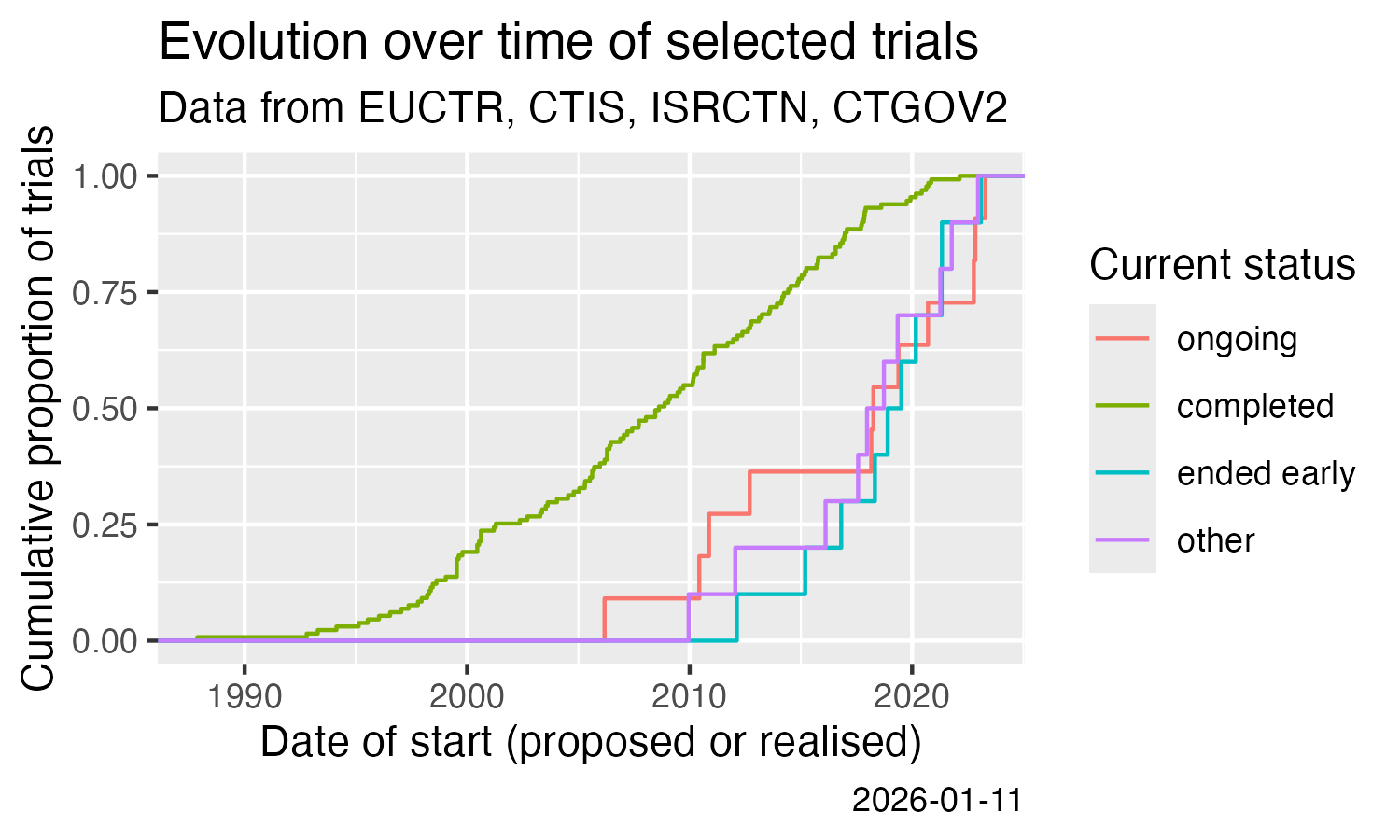
Show cumulative start of trials over time.
# use helper library
library(dplyr)
library(magrittr)
library(tibble)
# get names of all fields / variables in the collaction
length(dbFindFields(".*", con = db))
# [1] 3014
dbFindFields("(start.*date)|(date.*decision)", con = db)
# Using cache of fields.
# - Get trial data
result <- dbGetFieldsIntoDf(
fields = c(
"ctrname",
"record_last_import",
# CTGOV
"start_date",
"overall_status",
# CTGOV2
"protocolSection.statusModule.startDateStruct.date",
"protocolSection.statusModule.overallStatus",
# EUCTR
"n_date_of_competent_authority_decision",
"trialInformation.recruitmentStartDate", # needs above: 'euctrresults = TRUE'
"p_end_of_trial_status",
# ISRCTN
"trialDesign.overallStartDate",
"trialDesign.overallEndDate",
# CTIS
"authorizedPartI.trialDetails.trialInformation.trialDuration.estimatedRecruitmentStartDate",
"ctStatus"
),
con = db
)
# - Deduplicate trials and obtain unique identifiers
# for trials that have records in several registers
# - Calculate trial start date
# - Calculate simple status for ISRCTN
result %<>%
filter(`_id` %in% dbFindIdsUniqueTrials(con = db)) %>%
rowwise() %>%
mutate(start = max(c_across(matches("(date.*decision)|(start.*date)")), na.rm = TRUE)) %>%
mutate(isrctnStatus = if_else(trialDesign.overallEndDate < record_last_import, "Ongoing", "Completed")) %>%
ungroup()
# Merge variables / fields from different registers with re-leveling:
statusValues <- list(
"ongoing" = c(
# EUCTR
"Recruiting", "Active", "Ongoing", "Temporarily Halted", "Restarted",
# CTGOV
"Active, not recruiting", "Enrolling by invitation", "Not yet recruiting",
"ACTIVE_NOT_RECRUITING",
# CTIS
"Ongoing, recruiting", "Ongoing, recruitment ended",
"Ongoing, not yet recruiting", "Authorised, not started"
),
"completed" = c("Completed", "COMPLETED"),
"other" = c("GB - no longer in EU/EEA", "Trial now transitioned",
"Withdrawn", "Suspended", "No longer available",
"Terminated", "TERMINATED", "Prematurely Ended")
)
result[["state"]] <- dfMergeVariablesRelevel(
df = result,
colnames = c(
"overall_status", "p_end_of_trial_status",
"ctStatus", "isrctnStatus"
),
levelslist = statusValues
)
# example plot
library(ggplot2)
ggplot(result) +
stat_ecdf(aes(x = start, colour = state))
ggsave(
filename = "man/figures/README-ctrdata_across_registers.png",
width = 5, height = 3, units = "in"
)- Result-related trial information
Analyse some simple result details (see this vignette for more examples):
# Get all records that have values in any of the specified fields:
result <- dbGetFieldsIntoDf(
fields = c(
"clinical_results.baseline.analyzed_list.analyzed.count_list.count",
"clinical_results.baseline.group_list.group",
"clinical_results.baseline.analyzed_list.analyzed.units",
"study_design_info.allocation",
"location"
),
con = db
)
# Transform all fields into long name - value format:
result <- dfTrials2Long(df = result)
# Total 9472 rows, 12 unique names of variables
# [1.] Get counts of subjects for all arms into data frame
# This count is in the group where either its title or its
# description starts with "Total":
nsubj <- dfName2Value(
df = result,
valuename = "clinical_results.baseline.analyzed_list.analyzed.count_list.count.value",
wherename = paste0(
"clinical_results.baseline.group_list.group.title|",
"clinical_results.baseline.group_list.group.description"
),
wherevalue = "^Total$"
)
# Returning values for 32 out of 55 trials
# [2.] Count number of sites:
nsite <- dfName2Value(
df = result,
# some ctgov records use location.name, others location.facility.name
valuename = "^location.*name$"
)
# Returning values for 32 out of 55 trials
nsite <- tapply(
X = nsite[["value"]],
INDEX = nsite[["_id"]],
FUN = length,
simplify = TRUE
)
nsite <- data.frame(
"_id" = names(nsite),
nsite,
check.names = FALSE,
stringsAsFactors = FALSE,
row.names = NULL
)
# [3.] Randomised?
ncon <- dfName2Value(
df = result,
valuename = "study_design_info.allocation"
)
# Returning values for 27 out of 55 trials
# [4.] Merge sets:
nset <- merge(nsubj, nsite, by = "_id")
nset <- merge(nset, ncon, by = "_id")
# [5.] Example plot:
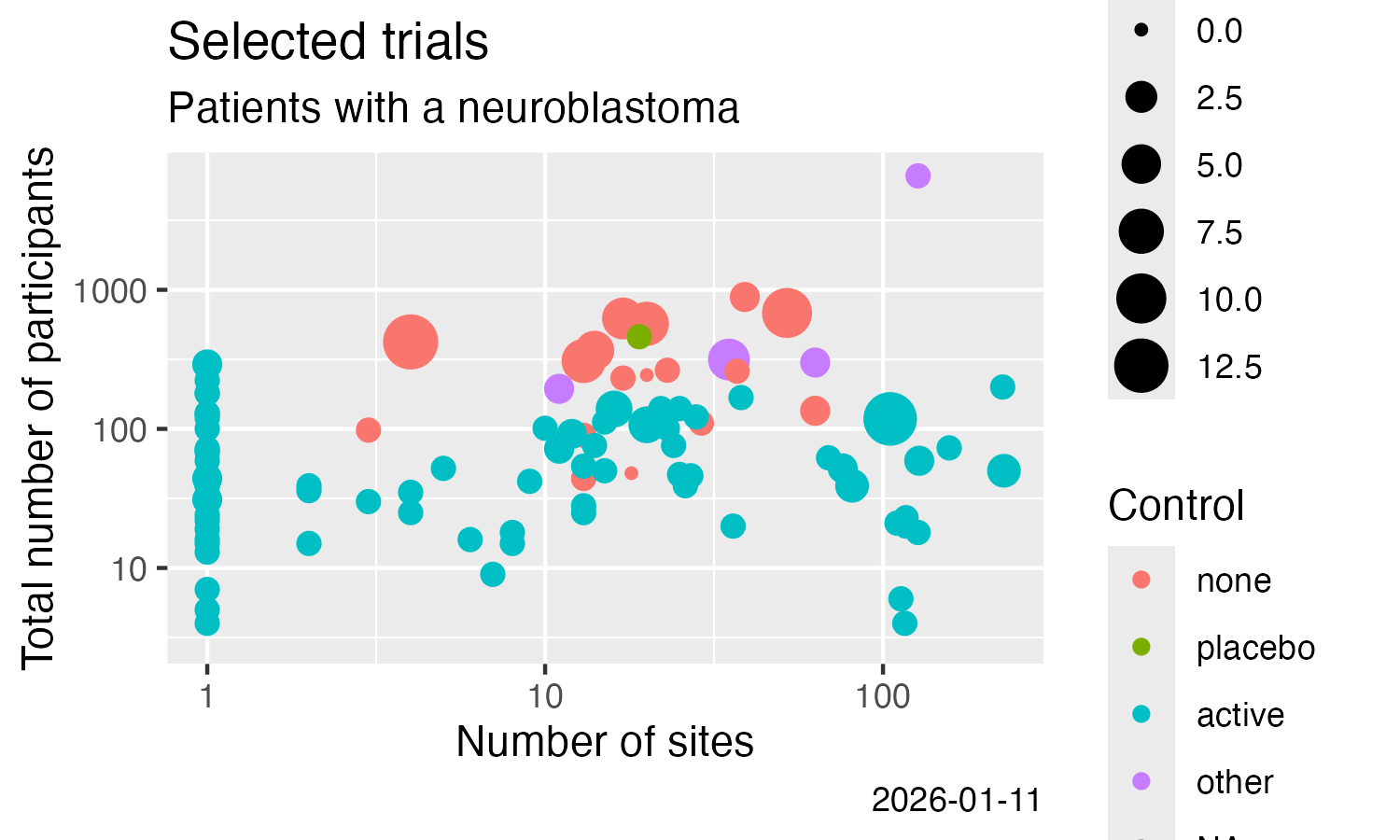
library(ggplot2)
ggplot(data = nset) +
labs(
title = "Neuroblastoma trials with results",
subtitle = "clinicaltrials.gov"
) +
geom_point(
mapping = aes(
x = nsite,
y = value.x,
colour = value.y == "Randomized"
)
) +
scale_x_log10() +
scale_y_log10() +
xlab("Number of sites") +
ylab("Total number of subjects") +
labs(colour = "Randomised?")
ggsave(
filename = "man/figures/README-ctrdata_results_neuroblastoma.png",
width = 5, height = 3, units = "in"
)- Download documents: retrieve protocols, statistical analysis plans and
other documents into the local folder
./files-.../
# EUCTR document files can be downloaded when results are requested
ctrLoadQueryIntoDb(
queryterm = "https://www.clinicaltrialsregister.eu/ctr-search/search?query=cancer&age=under-18&phase=phase-one",
euctrresults = TRUE,
documents.path = "./files-euctr/",
con = db
)
# * Found search query from EUCTR: query=cancer&age=under-18&phase=phase-one
# [...]
# Created directory ./files-euctr/
# Downloading trials...
# [...]
# = Imported or updated results for 102 trials
# = documents saved in './files-euctr'
# CTGOV files
ctrLoadQueryIntoDb(
queryterm = "cond=Neuroblastoma&type=Intr&recrs=e&phase=1&u_prot=Y&u_sap=Y&u_icf=Y",
register = "CTGOV",
documents.path = "./files-ctgov/",
con = db
)
# * Found search query from CTGOV: cond=Neuroblastoma&type=Intr&recrs=e&phase=1&u_prot=Y&u_sap=Y&u_icf=Y
# [...]
# Downloading documents into 'documents.path' = ./files-ctgov/
# - Created directory ./files-ctgov
# Applying 'documents.regexp' to 14 documents
# Downloading 10 documents:
# Download status: 10 done; 0 in progress. Total size: 38.10 Mb (100%)... done!
# Newly saved 10 document(s) for 7 trial(s); 0 document(s) for 0 trial(s) already existed
# CTIS files
ctrLoadQueryIntoDb(
queryterm = "https://euclinicaltrials.eu/app/#/search?ageGroupCode=2",
documents.path = "./files-ctis/",
con = db
)
# * Found search query from CTIS: ageGroupCode=2
# [...]
# * Downloading documents into 'documents.path' = ./files-ctis/
# - Created directory /Users/ralfherold/Daten/mak/r/emea/ctrdata/files-ctis
# - Getting ids of lists with document information
# - Downloading 564 lists with document information (estimate: 11.28 Mb)
# Download status: 564 done; 0 in progress. Total size: 7.39 Mb (100%)... done!
# - Processing document information in 564 lists
# - Creating subfolder for each trial
# - Applying 'documents.regexp' to 3132 documents
# - Downloading 278 missing documents
# Download status: 278 done; 0 in progress. Total size: 228.06 Mb (100%)... done!
# = Newly saved 255 document(s) for 29 trial(s) (latest versions only, deduplicated
# if e.g. in application and authorised part); 0 document(s) for 0 trial(s) already existed suppressMessages(tinytest::test_all())
# test_ctrdata_ctrfindactivesubstance.R 4 tests OK 1.7s
# test_ctrdata_mongo_local_ctgov.R 37 tests OK 59.6s
# test_ctrdata_mongo_local_ctis.R 19 tests OK 1.4s
# test_ctrdata_mongo_local_euctr.R 75 tests OK 2.3s
# test_ctrdata_mongo_local_isrctn.R 30 tests OK 7.4s
# test_ctrdata_other_functions.R 50 tests OK 0.6s
# test_ctrdata_sqlite_ctgov.R 37 tests OK 3.0s
# test_ctrdata_sqlite_ctis.R 19 tests OK 2.4s
# test_ctrdata_sqlite_euctr.R 75 tests OK 7.0s
# test_ctrdata_sqlite_isrctn.R 30 tests OK 9.7s
# All ok, 376 results (17m 25.3s)-
Canonical definitions, filters, calculations are in the works (since August 2023) for data mangling and analyses across registers, e.g. to define study population, identify interventional trials, calculate study duration; public collaboration on these canonical scripts will speed up harmonising analyses.
-
Authentication, expected to be required by CTGOV2; specifications not yet known (work not yet started).
-
Explore further registers such as ICTRP (authentication needed), JPRN, jRCT, UMIN-CTR, ChiCTR (exploration is continually ongoing; added value, terms and conditions for programmatic access vary; no clear roadmap is established yet).
-
Retrieve previous versions of protocol- or results-related information (work not yet started). The challenge is that, apparently, initial versions cannot be queried and historical versions can only be retrieved one-by-one and not in structured format.
-
Merge results-related fields retrieved from different registers, such as corresponding endpoints (work not yet started). The challenge is the incomplete congruency and different structure of data fields.
-
Data providers and curators of the clinical trial registers. Please review and respect their copyrights and terms and conditions, see
ctrOpenSearchPagesInBrowser(copyright = TRUE). -
Package
ctrdatahas been made possible building on the work done for R, clipr. curl, dplyr, duckdb, httr, jqr, jsonlite, lubridate, mongolite, nodbi, RPostgres, RSQLite, rvest, stringi and xml2.
-
Please file issues and bugs here. Also check out how to handle some of the closed issues, e.g. on C stack usage too close to the limit and on a SSL certificate problem: unable to get local issuer certificate
-
Information in trial registers may not be fully correct; see for example this publication on CTGOV.
-
No attempts were made to harmonise field names between registers (nevertheless,
dfMergeVariablesRelevel()can be used to merge and map several variables / fields into one).