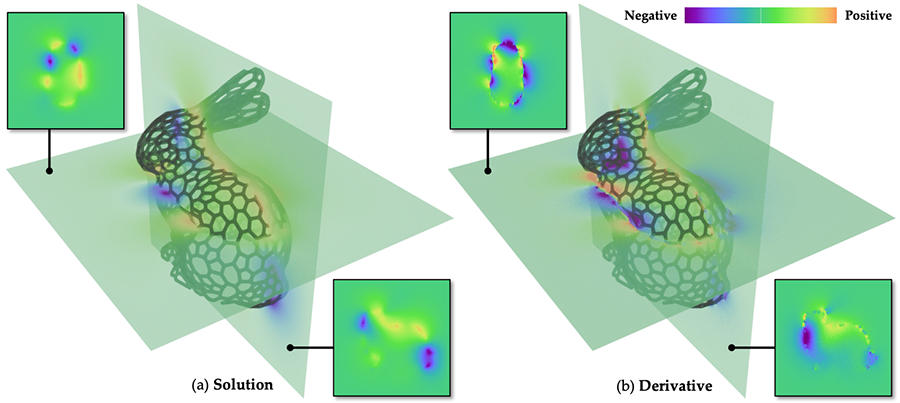
This repository contains the code for reproducing the results from the paper "A Differential Monte Carlo Solver For the Poisson Equation", Zihan Yu, Lifan Wu, Zhiqian Zhou, and Shuang Zhao, 2024.
The code is written in C++, CUDA, and Python. It has been tested on Ubuntu 20.04 with GCC 10.5.0, CUDA 12.2, and Python 3.11.
First, clone the repository:
git clone --recurse-submodules https://github.com/zihay/diff-wos.gitIf you are using conda, you can create a new environment with the following command:
conda env create -f environment.ymlThen, activate the environment and install the dependencies:
conda activate .condaFinally, compile the project:
mkdir build
cd build
CC=gcc-10 CXX=g++-10 cmake ..
cmake --build . --config ReleaseThe Python bindings will be compiled in the build/python directory. You can add this to your PYTHONPATH. If you're using vscode, the cloned .vscode folder should automatically add the path for you.
Important: Make sure the project, drjit, and mitsuba are compiled with identical compiler and settings for binary compatibility. If you've used pip to install pre-built versions of drjit and mitsuba, compile this project using GCC 10 for binary compatibility since pre-built packages were compiled with it. The compiler used for pre-built packages can be found here. Failing to do so may lead to type-mismatch errors due to binary incompatibility.
diff_solve directory contains the differentiable PDE solve examples. You can replicate the results by directly running the scripts in that directory. By default, the scripts will run the primal Walk-on-Spheres solver. To run the differential solvers, uncomment the corresponding lines in the scripts.
inv_solve directory contains the inverse PDE solve examples. You can replicate the results by directly running the scripts in that directory. By default, the scripts will run the optimization with our method. To run the baseline methods, uncomment the corresponding lines in the scripts.
The ablation study evaluates the performance of four normal-derivative estimators and is located in the ablations directory. You can replicate the results by directly running the scripts in that directory.
@inproceedings{10.1145/3641519.3657460,
author = {Yu, Zihan and Wu, Lifan and Zhou, Zhiqian and Zhao, Shuang},
title = {A Differential Monte Carlo Solver For the Poisson Equation},
year = {2024},
address = {New York, NY, USA},
doi = {10.1145/3641519.3657460},
booktitle = {ACM SIGGRAPH 2024 Conference Papers},
}