YADA is an innovative biological deconvolution algorithm developed by the author and a collaborator as part of doctoral research in the Systems Biomedicine Lab under the supervision of Professor Efroni. The purpose of YADA is to estimate the proportions of distinct cell types within complex, heterogeneous gene expression samples.
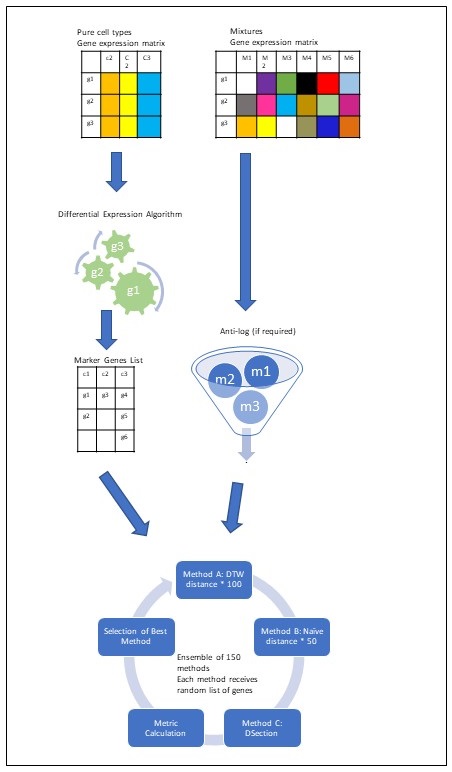
The fundamental premise behind YADA is that the transcriptomic signatures of pure cell populations can be leveraged to deconvolute mixed expression profiles and quantify the relative abundance of each constituent cell type. By analyzing gene expression patterns, deconvolution algorithms computationally unravel the complexities inherent to these cellular mixtures.
YADA implements a robust approach to perform this deconvolution task, accurately estimating immune and other cell type fractions from bulk transcriptomic data. It represents a novel contribution stemming from the author's doctoral studies focused on advancing computational methods for dissecting intricate systems-level biomedical data. Under the guidance of Professor Efroni, the authors were able to design, validate, and optimize YADA to address a crucial need in the field of computational immunology.
YADA implements two approaches for deconvolution:
- Marker-based deconvolution using curated gene signatures representative of each cell type.
- Reference-based deconvolution utilizing a complete gene expression matrix from pure cell populations.
The key features and innovations of YADA include:
- High performance: YADA achieves state-of-the-art accuracy on benchmark datasets, as evidenced by top results in a recent challenge.
- Flexibility: Works with either marker genes or full reference profiles.
- Broad applicability: Core algorithm supports gene expression data from various sequencing platforms.
- Computational efficiency: Optimized parallel implementation allows rapid deconvolution of large datasets.
- User-friendly Python API: One of the few deconvolution libraries natively implemented in Python.
pip install yada_deconv or Clone this repository using the git clone command.
The training dataset for YADA comprises benchmark datasets available in the "data" folder. This comprehensive collection includes data from publicly available sources as well as synthetically generated datasets created by the authors. These datasets were utilized for training and validation purposes during the DREAM challenge, a community-based deconvolution benchmarking effort.
The input data for YADA consists of two files:
pure.csv: A gene expression matrix for purified cell populations, with dimensions(n_genes) x (k_cell_types). For marker-based deconvolution, this file should contain only gene symbols. Refer to the sample notebook for formatting details.mix.csv: A gene expression matrix for mixed cell samples, with dimensions(n_genes) x (m_mixtures). The first row must contain mixture labels.
Additional guidelines for input files:
- Gene symbols should be provided in the first column for both files.
- It is acceptable for some genes to be missing in either the pure or mixed file.
- Expression data should be in non-log scale. If the maximum expression value is <50, an anti-log transformation is automatically applied.
- YADA performs internal marker gene selection; therefore, not all provided signature genes may be utilized.
By following these specifications, users can ensure their input data is properly formatted for YADA to perform accurate deconvolution.
from YADA import *
#Marker gene list.
pure_file_path = '../data/xCell/pure.csv'
#This is the mixture file in the format: columns: mix1, mix2, ..., rows: gene names.
mix_file_path = '..data/xCell/mix.csv'
result = run_yada(pure, mix)
- Using marker gene list
- Using a complete reference matrix
- YADA challenge result
- YADA simulation running
We welcome contributions! Please see our Contributing Guidelines for more information on how to get involved.
YADA is available under the MIT license. See the LICENSE file for more details.
For questions, issues, or feature requests, please open an issue on our GitHub repository. For additional support, contact: zurkin at yahoo dot com.
If you use YADA in your research, please cite our paper: Livne Dani, Snir Tom, Efroni Sol, YADA - Reference Free Deconvolution of RNA Sequencing Data, Current Bioinformatics; Volume 19, Issue , Year 2024, e050624230728. DOI: 10.2174/0115748936304034240405034414, https://www.eurekaselect.com/article/140845
We thank the scientific community for their valuable feedback and contributions to this project.
- Matrix decomposition https://en.wikipedia.org/wiki/Matrix_decomposition
- Collection of papers https://github.com/changwn/Deconvolution_paper