A Deep Learning-Based Model for Quantifying Transposable Elements in Single-Cell Sequencing Data
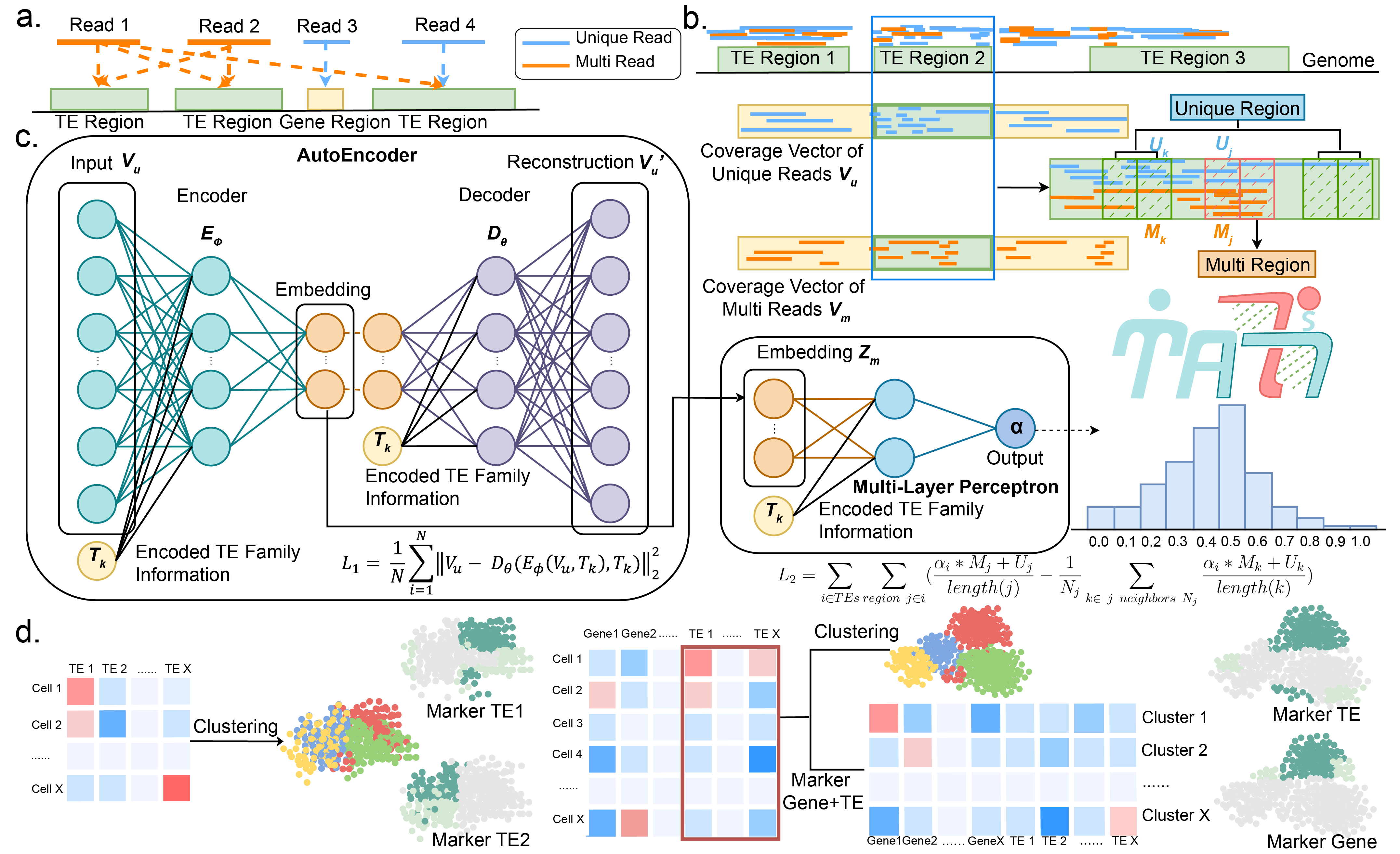
MATES is a specialized tool designed for precise quantification of transposable elements (TEs) in various single-cell datasets. The workflow consists of multiple stages to ensure accurate results. In the initial phase, raw reads are mapped to the reference genome, differentiating between unique-mapping and multi-mapping reads associated with TE loci. Unique-mapping reads create coverage vectors (Vu), while multi-mapping reads remain associated with Vm vectors, both capturing read distribution around TEs. TEs are then divided into bins, either unique-dominant (U) or multi-dominant (M), based on read proportion. An autoEncoder model is employed to create latent embeddings (Zm) capturing local read context and is combined with TE family information (Tk). In the subsequent stage, the obtained embeddings are used to jointly estimate the multi-mapping ratio (αi) via a multilayer perceptron. Training the model involves a global loss (L1 and L2) comprising reconstruction loss and read coverage continuity. Trained to predict multi-mapping ratios, the model counts reads in TE regions, enabling probabilistic TE quantification at the single-cell level. MATES enhances cell clustering and biomarker identification by integrating TE quantification with gene expression methods.With the burgeoning field of single-cell sequencing data, the potential for in-depth TE quantification and analysis is enormous, opening avenues to gain invaluable insights into the molecular mechanisms underpinning various human diseases. MATES furnishes a powerful tool for accurately quantifying and investigating TEs at specific loci and single-cell level, thereby significantly enriching our understanding of complex biological processes. This opens a new dimension for genomics and cell biology research and holds promise for potential therapeutic breakthroughs.
- Version 0.1.5: Improve the efficiency of splitting BAM files and counting TEs reads.
- Version 0.1.4: Enhanced the build_reference.py script and the tutorial to build reference genome for species other than Human and Mouse.
MATES is actively under development; please feel free to reach out if you encounter any issues.
To install MATES, you can run the following command:
# Clone the MATES repository
git clone https://github.com/mcgilldinglab/MATES.git
# Create a new environment
conda create -n mates_env python=3.9
conda activate mates_env
# Install required packages
conda install -c bioconda samtools -y
conda install -c bioconda bedtools -y
# Install MATES
cd MATES
pip install .
# Add environment to Jupyter Notebook
conda install ipykernel
python -m ipykernel install --user --name=mates_envInstallation should take only a few minutes. Verify that MATES is correctly installed by running in python:
import MATESInteractive MATES web server: https://mates.cellcycle.org.
We have supported automatic human and mouse TE/Gene reference genome creating using python build_reference.py --species Human/Mouse. For Arabidopsis thaliana and Drosophila melanogaster, please visit the shared folder for GTF file, RepeatMaskers file, and example script to create their TE/Gene reference genome. For other species, please refer to the tutorial of building TE and Gene reference genome.
From loading data to downstream analysis. Please refer to Example Section for deatils.
- MATES pipeline on Smart-seq2 scRNA and 10X scRNA/scATAC/Multi-Omics data
- MATES pipeline on 10X Visium data
- MATES pipeline on Long Reads data
- MATES downstream analysis on Smart-seq2 scRNA data (TE only)
- MATES downstream analysis on Smart-seq2 scRNA data (Gene+TE)
The MATES contains six modules.
import MATES
from MATES import bam_processor
from MATES import data_processor
from MATES import MATES_model
from MATES import TE_quantifier
from MATES import TE_quantifier_LongRead
from MATES import TE_quantifier_Intronic- bam_processor The bam_processor module efficiently manages input BAM files by partitioning them into sub-BAM files for individual cells, distinguishing unique mapping from multi mapping reads. It also constructs TE-specific coverage vectors, shedding light on read distributions around TE instances at the single-cell level, enabling accurate TE quantification and comprehensive cellular characterization.
In the MATES v0.1.5, we released a new function bam_processor.split_count_10X_data() to speed up the preprocessing step for 10X data.
bam_processor.split_count_10X_data(TE_mode,data_mode, sample_list_file, bam_path_file, bc_path_file, bc_ind='CB', ref_path = 'Default')
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene (for intronic, refer to below section)
## data_mode : <str> 10X, This function currently only support 10X data.
## sample_list_file : <str> path to file conatins sample IDs
## bam_path_file : <str> path to file conatins matching bam file address of sample in sample list
## bc_path_file(optional) : <str> path to file contains matching barcodes list address of sample in sample list
## bc_ind:<str> barcode field indicator in bam files, e.g. CB/CR...
## ref_path(optional): <str> TE reference bed file. Only needed for self generated reference, provide path to reference. By default, exclusive have reference 'TE_nooverlap.bed' and inclusive have reference 'TE_full.bed'.bam_processor.split_bam_files(data_mode, threads_num, sample_list_file, bam_path_file,bc_ind = None, bc_path_file=None)
# Parameters
## data_mode : <str> 10X or Smart_seq, we encourage you to use bam_processor.split_count_10X_data() for 10X data.
## threads_num : <int>
## sample_list_file : <str> path to file conatins sample IDs
## bam_path_file : <str> path to file conatins matching bam file address of sample in sample list
## bc_ind:<str> barcode field indicator in bam files, e.g. CB/CR...
## bc_path_file(optional) : <str> path to file contains matching barcodes list address of sample in sample listbam_processor.count_coverage_vec(TE_mode, data_mode, threads_num, sample_list_file, ref_path = "Default", bc_path_file=None)
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene (for intronic, refer to below section)
## data_mode : <str> 10X or Smart_seq, we encourage you to use bam_processor.split_count_10X_data() for 10X data.
## threads_num : <int>
## sample_list_file : <str> path to file conatins sample IDs
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default, exclusive have reference 'TE_nooverlap.csv' and inclusive have reference 'TE_full.csv'.
## bc_path_file(optional) : <str> only needed for 10X data, path to file contains matching barcodes list address of sample in sample listIf you want to perform TE quantification on Long Reads data, you can use bam_processor.split_bam_files based on your sequencing plantform. Instead of using bam_processor.count_coverage_vec, use below function:
For simplicity, in data_mode, we use 10X to indicating data using barcodes to distinguish data, i.e. you may have a barcode file to seperating the data in the bam file or Smart_seq to indicating data do not use barcodes to distinguish data, i.e. one bam file per cell.
bam_processor.count_long_reads(TE_mode, data_mode, threads_num, sample_list_file, bam_dir, ref_path = "Default", bc_path_file=None):
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene
## data_mode : <str> 10X or Smart_seq
## threads_num : <int>
## sample_list_file : <str> path to file conatins sample IDs
## bam_dir: <str> path to director conatins sample bam files
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default, exclusive have reference 'TE_nooverlap.csv' and inclusive have reference 'TE_full.csv'.
## bc_path_file(optional) : <str> only needed for data using barcodes to distinguish data, path to file contains matching barcodes list address of sample in sample list- data_processor The data_processor module assists in computing Unique and Multi Regions, generating training samples, and summarizing the expression of multi-mapping reads for prediction.
data_processor.calculate_UM_region(TE_mode, data_mode, sample_list_file, bin_size=5, proportion=80, ref_path = "Default", bc_path_file=None)
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default, exclusive have reference 'TE_nooverlap.csv' and inclusive have reference 'TE_full.csv'.
## bc_path_file(optional) : <str> only needed for 10X data, path to file contains matching barcodes list address of sample in sample listdata_processor.generate_training_sample(data_mode, sample_list_file, bin_size, proportion)
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80data_processor.generate_prediction_sample(TE_mode, data_mode,sample_list_file, bin_size, proportion, ref_path = "Default",bc_path_file=None)
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default, exclusive have reference 'TE_nooverlap.csv' and inclusive have reference 'TE_full.csv'.
## bc_path_file(optional) : <str> only needed for 10X data, path to file contains matching barcodes list address of sample in sample list- MATES_model The MATES_model module serves as the core of the MATES framework, encompassing both training and prediction functions. It is responsible for training a neural network model to accurately predict multi-mapping rates of transposable element (TE) instances based on their read coverage vectors.
MATES_model.train(data_mode, sample_list_file, bin_size = 5, proportion = 80, BATCH_SIZE= 4096, AE_LR = 1e-4, MLP_LR = 1e-6, AE_EPOCHS = 200, MLP_EPOCHS = 200, USE_GPU= True)
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80
## BATCH_SIZE : <int> default = 4096
## AE_LR : <int> learning rate of AutoEncoder, default = 1e-4
## MLP_LR : <int> learning rate of MLP, default = 1e-6
## AE_EPOCHS : <int> training epochs for AutoEncoder, default = 200
## MLP_EPOCHS : <int> training epochs for MLP, default = 200
## USE_GPU : <bool> whether use GU to train the model, default = TrueMATES_model.prediction(TE_mode, data_mode, sample_list_file, bin_size = 5, proportion = 80, AE_trained_epochs =200, MLP_trained_epochs=200, USE_GPU= True)
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80
## AE_EPOCHS : <int> training epochs for AutoEncoder, default = 200
## MLP_EPOCHS : <int> training epochs for MLP, default = 200
## USE_GPU : <bool> whether use GU to train the model, default = Truet- TE_quantifier TE_quantifier module facilitates the quantification of TE expression from unique mapping reads and organizes the generation of finalized TE matrix output files.
TE_quantifier.unique_TE_MTX(TE_mode, data_mode, sample_list_file, threads_num, bc_path_file=None)
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## threads_num : <int>
## bc_path_file(optional) : <str> only needed for 10X data, path to file contains matching barcodes list address of sample in sample listTE_quantifier.finalize_TE_MTX(data_mode, sample_list_file=None)
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file(optional) : <str> only needed for 10X data, path to file conatins sample IDs- TE_quantifier_LongRead TE_quantifier_LongRead module facilitates the quantification of TE expression from unique mapping reads at locus level for Long Read data.
TE_quantifier_LongRead.quantify_locus_TE_MTX(TE_mode, data_mode, sample_list_file)
# Parameters
## TE_mode : <str> exclusive or inclusive, represents whether remove TE instances have overlap with gene
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## long_read : <bool> whether you're quantifying long read data- TE_quantifier_Intronic TE_quantifier_Intronic module facilitates the quantification of TE expression in Intronic TEs.
implement_velocyto(data_mode, threads_num, sample_list_file, bam_path_file, gtf_path, bc_path_file=None)
# Parameters
## data_mode : <str> 10X or Smart_seq
## threads_num : <int>
## sample_list_file : <str> path to file conatins sample IDs
## bam_path_file : <str> path to file conatins matching bam file address of sample in sample list
## gtf_path : <str> path to the gene gtf file, this is mandatory to implement velocyto
## bc_path_file(optional) : <str> path to file contains matching barcodes list address of sample in sample listparse_velocyto_output(data_mode, threads_num, sample_list_file)
# Parameters
## data_mode : <str> 10X or Smart_seq
## threads_num : <int> threads to use (CPU number)
## sample_list_file : <str> path to file conatins sample IDscount_unspliced_reads(data_mode, threads_num, sample_list_file, ref_path='Default')
# Parameters
## data_mode : <str> 10X or Smart_seq
## threads_num : <int>
## sample_list_file : <str> path to file conatins sample IDs
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default TE reference is of name 'TE_intronic.csv'. count_intornic_coverage_vec(data_mode, threads_num, sample_list_file, ref_path = 'Default',bc_path_file=None)
# Parameters
## data_mode : <str> 10X or Smart_seq
## threads_num : <int> threads to use (CPU number)
## sample_list_file : <str> path to file conatins sample IDs
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default TE reference is of name 'TE_intronic.csv'.
## bc_path_file(optional) : <str> path to file contains matching barcodes list address of sample in sample listgenerate_prediction_sample(data_mode, sample_list_file, bin_size, proportion, ref_path = 'Default', bc_path_file=None)
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default TE reference is of name 'TE_intronic.csv'.
## bc_path_file(optional) : <str> path to file contains matching barcodes list address of sample in sample listquantify_U_TE_MTX(data_mode, sample_list_file)
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDsquantify_M_TE_MTX(data_mode, sample_list_file, bin_size=5, proportion=80, AE_trained_epochs=200, MLP_trained_epochs=200, USE_GPU= True, ref_path = 'Default')
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## bin_size : <int> size of U/M Region, default = 5
## proportion : <int> proportion of dominated unique reads in U Region / multi reads in M Region, default = 80
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default TE reference is of name 'TE_intronic.csv'.
## AE_EPOCHS : <int> training epochs for AutoEncoder, default = 200
## MLP_EPOCHS : <int> training epochs for MLP, default = 200
## USE_GPU : <bool> whether use GU to train the model, default = Truetcorrect_intronic_TE(data_mode, sample_list_file, ref_path = 'Default')
# Parameters
## data_mode : <str> 10X or Smart_seq
## sample_list_file : <str> path to file conatins sample IDs
## ref_path(optional): <str> only needed for self generated reference, provide path to reference. By default TE reference is of name 'TE_intronic.csv'.