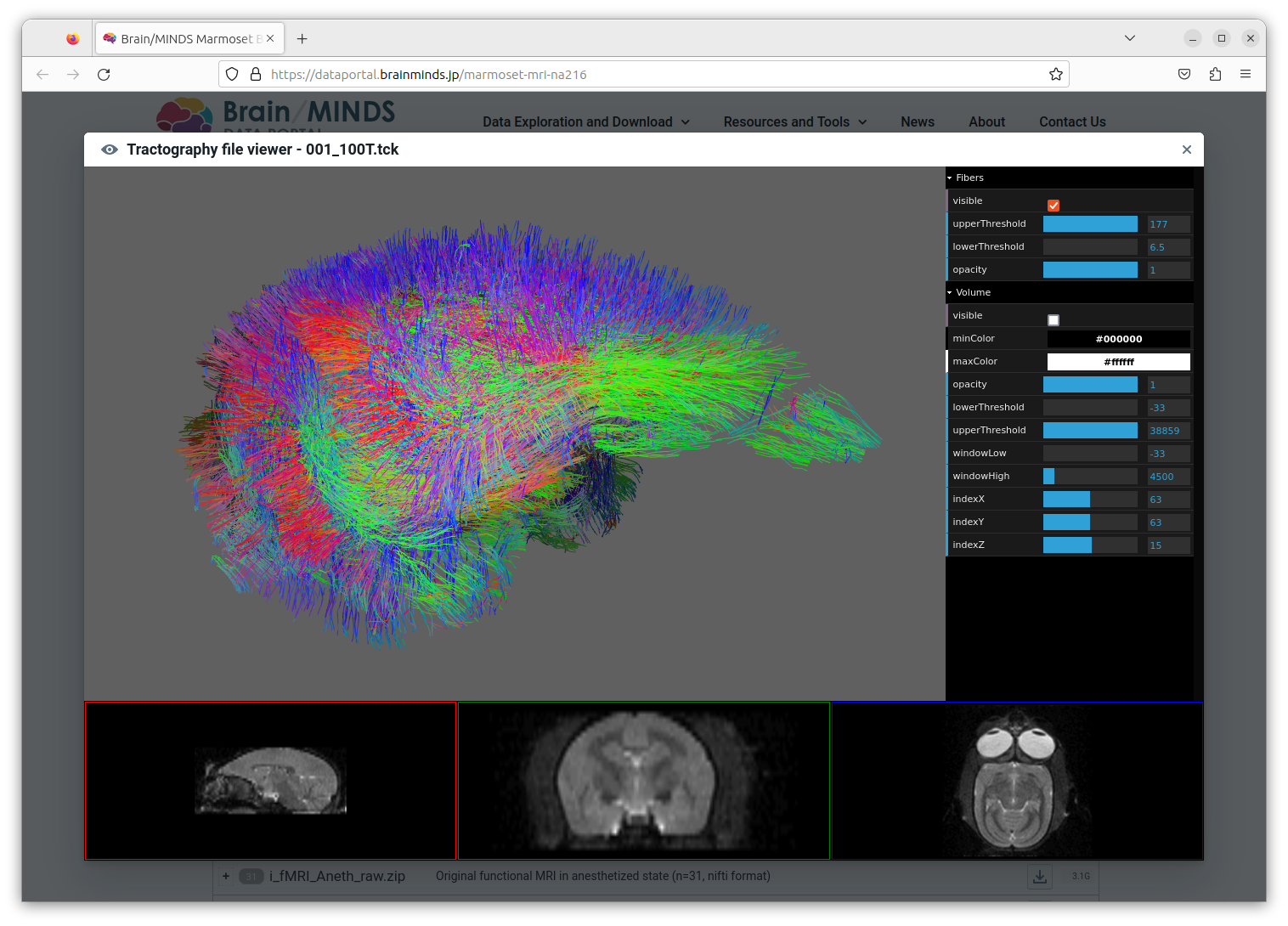
On the dataportal (see NA216 dataset), tractography data can be inspected with an online previewer leaveraging XTk javascript library. In practice, the rendered data often looks like a dense ball of fibers, and that's not so easy to explore the fibers details, especially for the inner regions which are obscured by outer regions.
Indeed, with Xtk it's only possible:
- to change the opacity of all fibers (of a trk file) at once,
- to hide some of them depending on their length (by setting min and max fibers length filter values).
It would be good to somehow be able to only view fibers inside a user-defined region of the space. (Tanaka-sensei proposal "three othogonal guide 'planes' of some thickness. Center and thickness can be specified in the UI with sliders").
Ideally by continuing using Xtk which is currently used by the online preview.
Extending tractography viewer to only display fibers in a user-defined volume (rectangular cuboid aligned to axis).
It means cropping the fiber rendering outside of the user defined volume, as figured in the schema below:
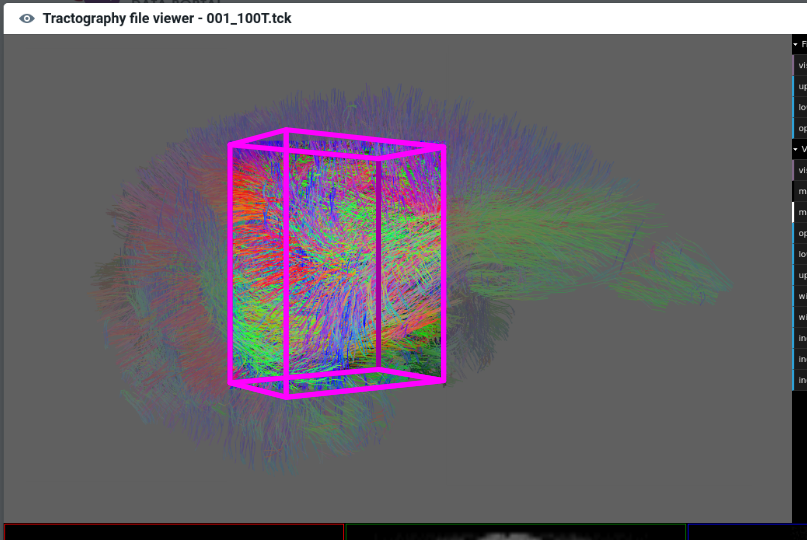
-
XTk is webGL powered; It defines a couple of generic shaders (in
shader.js) that can handle rendering of NifTI volumes, tractography fibers and more.
(see Webgl fundamentals for explanations about Webgl shaders) -
The shaders already includes mechanisms to adjust rendering of loaded objects depending on the value of user-defined criteria. For example, in case of tractography data, some individual fibers may be masked depending on their overall length.
-
Fiber files in TrackVis format (
.trk) are parsed byparserTRK.js. Each fiber segment is assigned a few scalars, including the overall length of the fiber they belong to. This enables the shader to discard the rendering of segments depending the length of their fiber.
(For fibers, WebGLRendering mode isLINES, to draw lines segment between pairs of vertices. GLSL discard keyword is used the shaders to hide fiber segments.)
In a first approach, we could probably obtain the desired effect of volume cropping by simply preventing the rendering of fiber segments that extend outside the user-defined volume of interest (similar logic as length based fiber masking described above). This simple approach is implemented first because it only requires limited changes in the shaders GLSL code (Xtk was written in GLSL ES 1.0); However, it has some limitations described below.
This would work if fibers are composed of many short-length line segments (which seems a reasonable assumption since in practice fibers tend not to have long straight stretches).
But, because in this first approximation the fibers would not be strictly cut at the volume edges (but rather at the boundaries of the neighbouring segment completely contained in the cuboid), the volume boundaries might not appear neat.
Moreover, we have to expect that when users define very thin volume (compared to segment length), the number of rendered fibers might become artificially very low.
We'll need to asses whether these artefacts are acceptable in the context of an online preview.
Basically :
- provide a UI to allow users to define their volume of interrest (cuboid) [use sliders with dat],
- send the cuboid definition to the shader [by means of newly defined
UNIFORMs], - extend vertex shader GLSL code to discard any segment extending outside of the cuboid (at least one of its vertice is outside).
✅ Here is a short clip showing the result of implementing solution #1
tracto-preview-volcropping-1.mp4
The first approach flaw is that it doesn't render at all segments that have some part outside of the cuboid volume.
Instead of discarding each of these segments as a whole, it is of course more natural to split these segments, discard the sub-segment outside of the volume while rendering the part inside.
It means redefining the coordinates of one or both vertices of the segment in the shader code, which is of course allowed.
But this requires much more changes in the code, and learn a bit more about shaders developement.
Although the vertex shader process one vertex at a time, we need both segment vertices coordinates to cut the segments at the cropping planes intersection, but it is not straightworward to obtain the other vertex coordinates in shader written in GLSL ES 1.0.
So we could take advantage of some functionnalities by switch to WebGL 2.0 (which is OK nowadays since more 90% of browsers support it), because the newer version of GLSL (ES 3.00) includes additions that are useful in our used case (such as built-in vertex shader variables gl_VertexID and others).
There some syntax changes in current shader code that need to be addressed to make it compatible with GLSL ES 3.00.
❌ The implementation of solution #2 could not be achieved during the time of the Hackathon 😓
-
demos/demo-before/contains a standalone demo similar to the current preview available on Brain/MINDS data portal (NA216 dataset). -
demos/demo-dev/contains the demo that runs on the Xtk version being developed (i.e. the one shipped as submodule within this repo) -
demos/demo-solution-1/contains a standalone demo of solution #1 (with Xtk bundle built at https://github.com/bm-hackathon-23/X/commit/4ba87e1e6f716ca42709323d0e1c1138faebd29a).
References:
- XTk Lesson #06: Connectivity http://lessons.goxtk.com/06/
- XTk Demo FiberAtlas http://demos.goxtk.com/brainfibers/
- git
npminstalled on your machine
git clone https://github.com/bm-hackathon-23/tracto-previewModern web-browsers don't allow loading local resources; Thus, a little http-server is needed for the demo to be able to load data.
cd tracto-preview/demos/
npm installDepending on which demo you want to run, change the directory parameter for http-server on the following command :
cd tracto-preview/
node_modules/http-server/bin/http-server demo-before -p 8000
4. Load http://127.0.0.1:8000/ in your web-browser.
Press [Ctrl]-c to stop the server.
cd tracto-preview/demos/demo-dev/X
git submodule init
git submodule updateRefer to Xtk's developer wiki section.
Xtk is shipped with specific python scripts to generate a deployable js bundle.
Because Xtk project has not been updated for a while, we had trouble to run building scripts with nowadays' version of python/java.
We can instead use a dedicated container just for this task.
cd tracto-preview/scripts
docker build -f Dockerfile.xtk-builder -t xtk-builder .
cd tracto-preview/
docker run -it --rm --user $(id -u $USER):$(id -g $USER) --name xtk-run-build -v `pwd`/demos/demo-dev/X:/usr/src/X -w /usr/src/X xtk-builder python utils/deps.py
cd tracto-preview/demos/
node_modules/http-server/bin/http-server /demo-dev -p 8000Load http://127.0.0.1:8000/ in your web-browser.
cd tracto-preview/
docker run -it --rm --user $(id -u $USER):$(id -g $USER) --name xtk-run-build -v `pwd`/demos/demo-dev/X:/usr/src/X -w /usr/src/X xtk-builder python utils/build.py